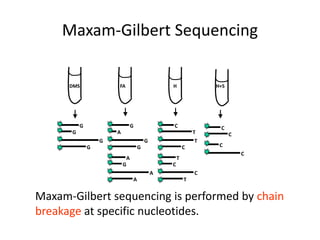
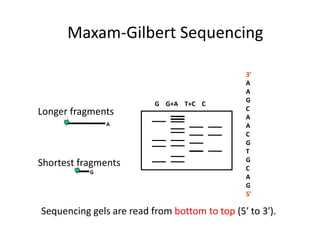
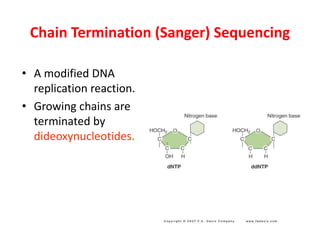
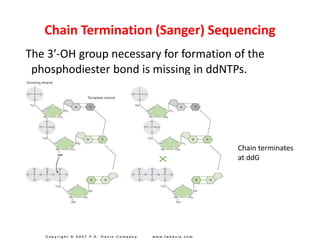
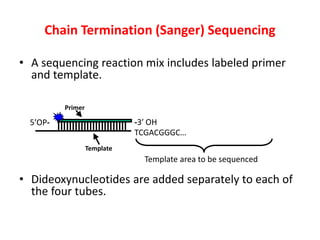
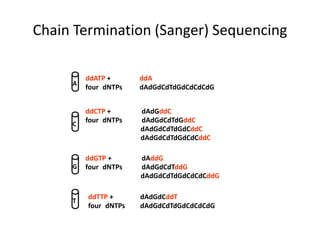
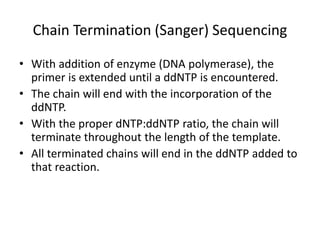
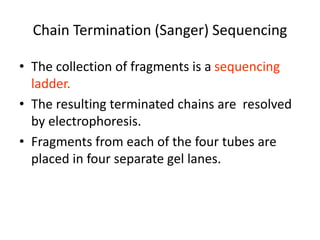
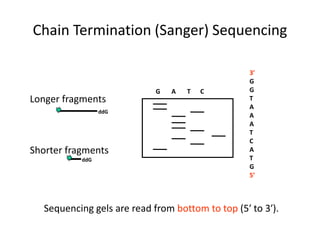
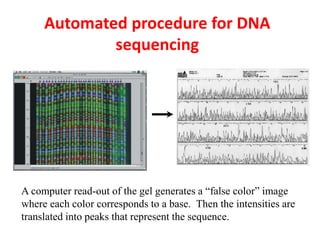
DNA sequencing methods determine the order of nucleotides in a DNA molecule. Maxam-Gilbert sequencing uses specific chemical agents to randomly cleave DNA fragments at individual nucleotide positions, generating labeled fragments that are separated by gel electrophoresis. Sanger sequencing uses DNA polymerase and modified nucleotides called dideoxynucleotides to terminate DNA replication in a sequence-specific manner, producing a collection of labeled fragments of different lengths that define the DNA sequence. Automated DNA sequencing now translates gel images into nucleotide sequences using fluorescent tags and detection of signal intensities.