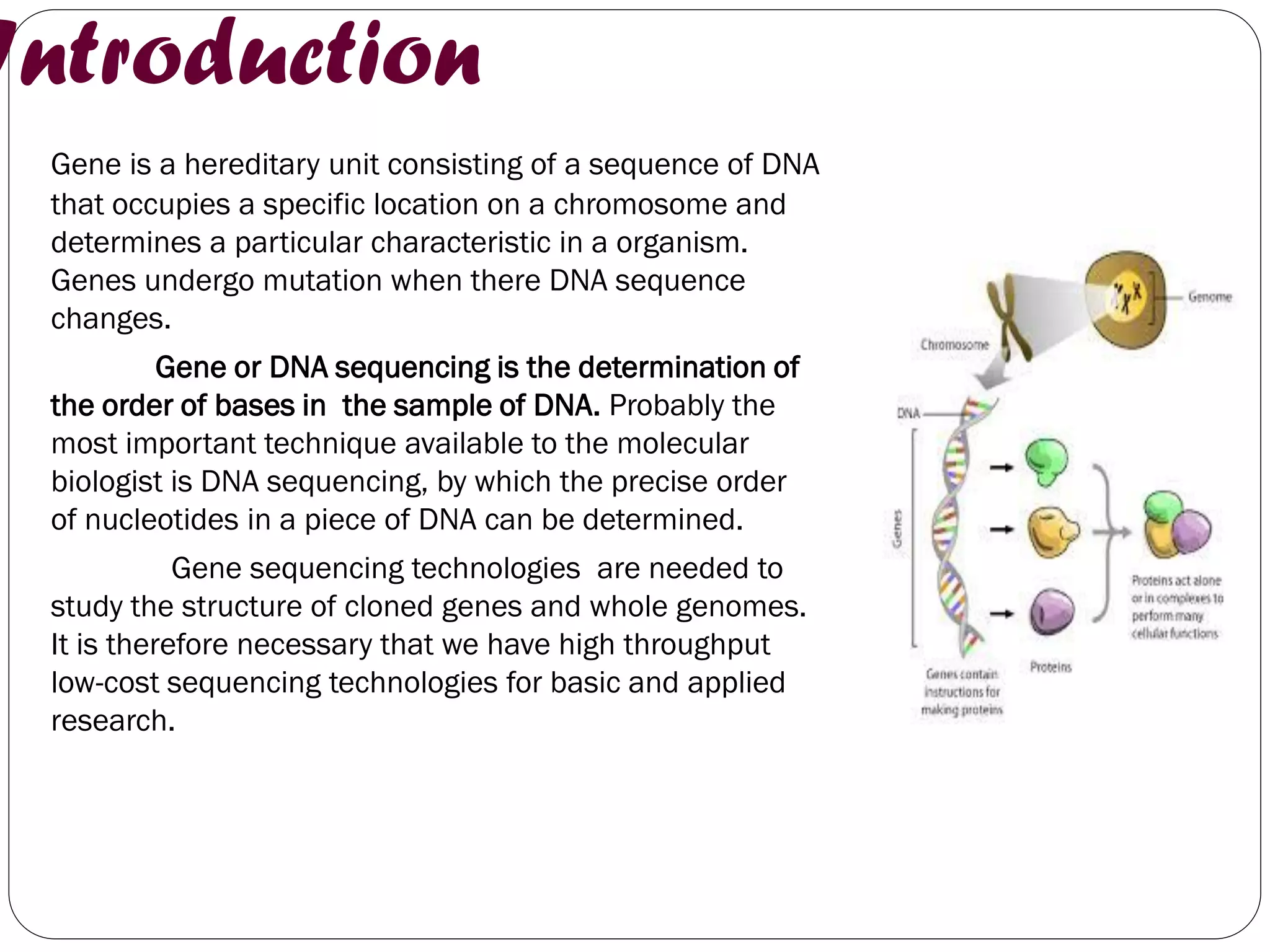
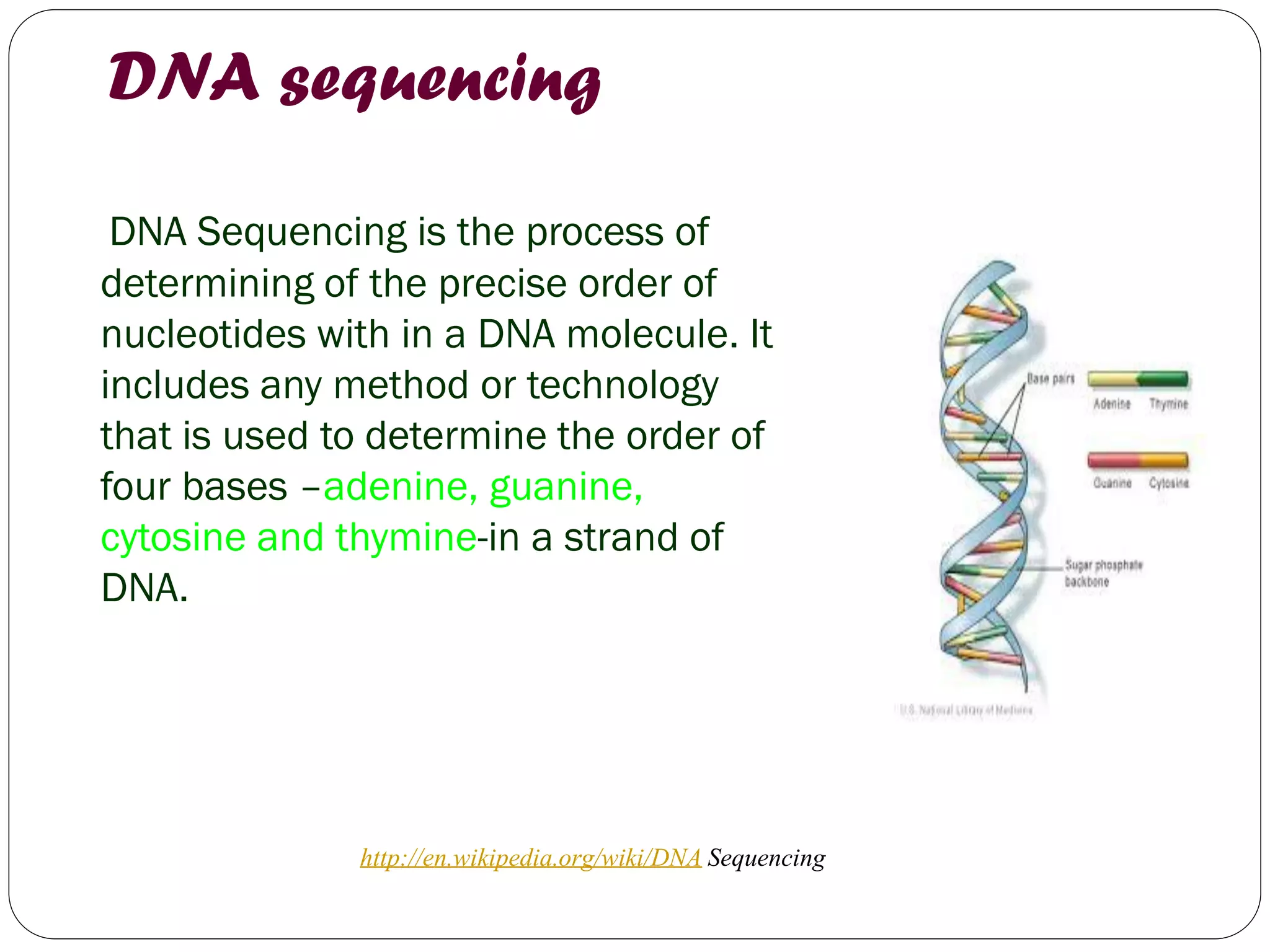
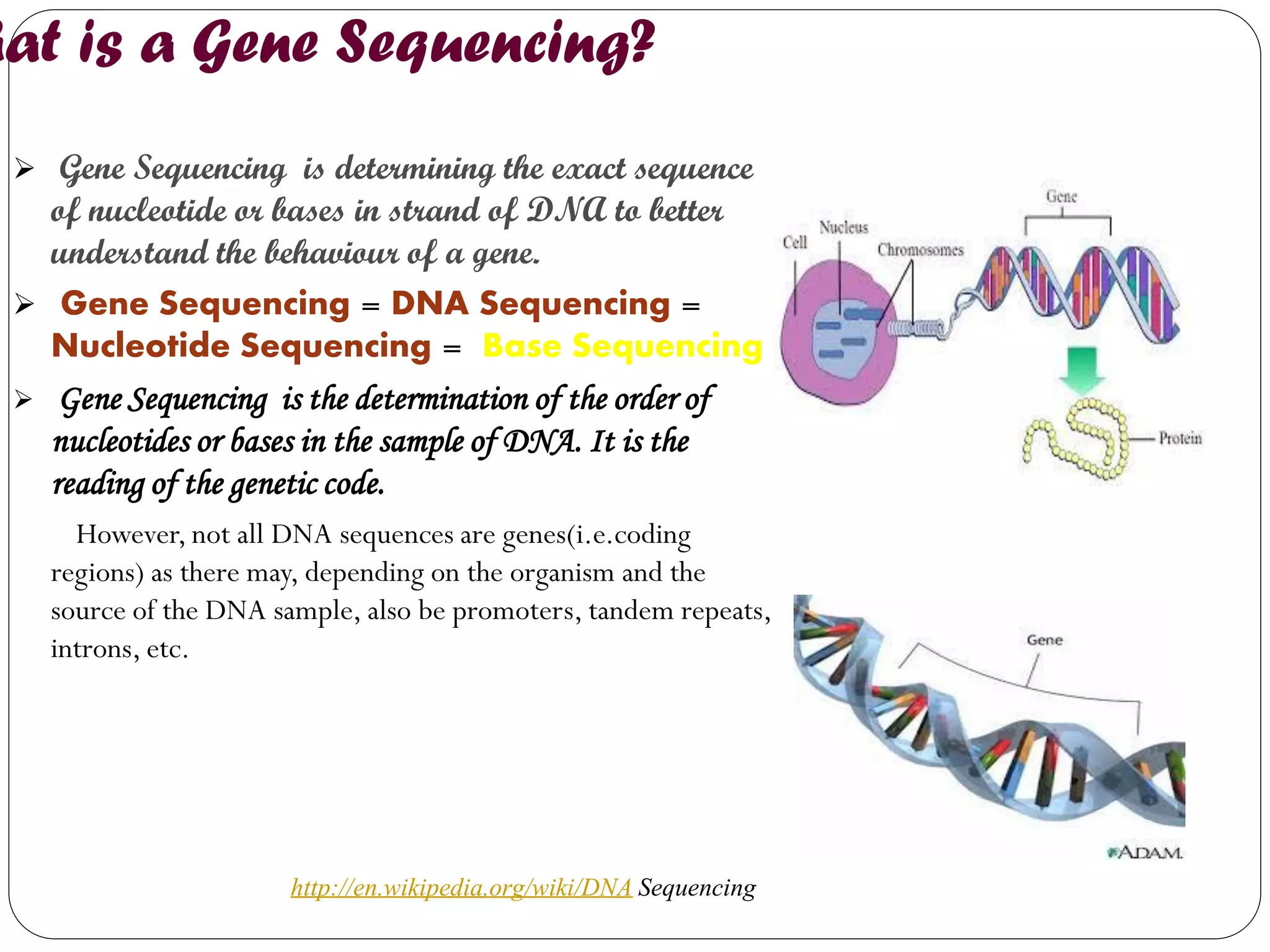
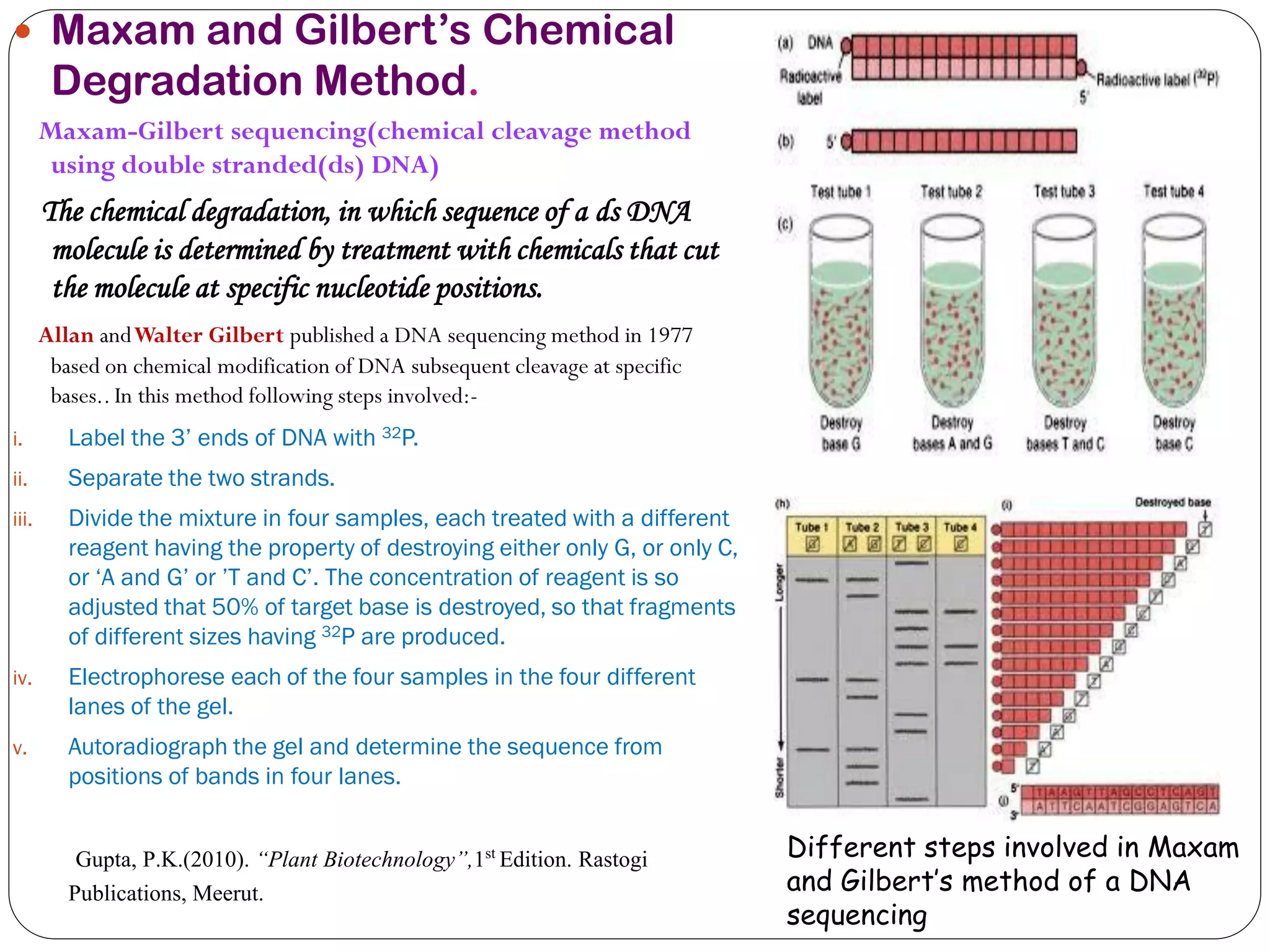
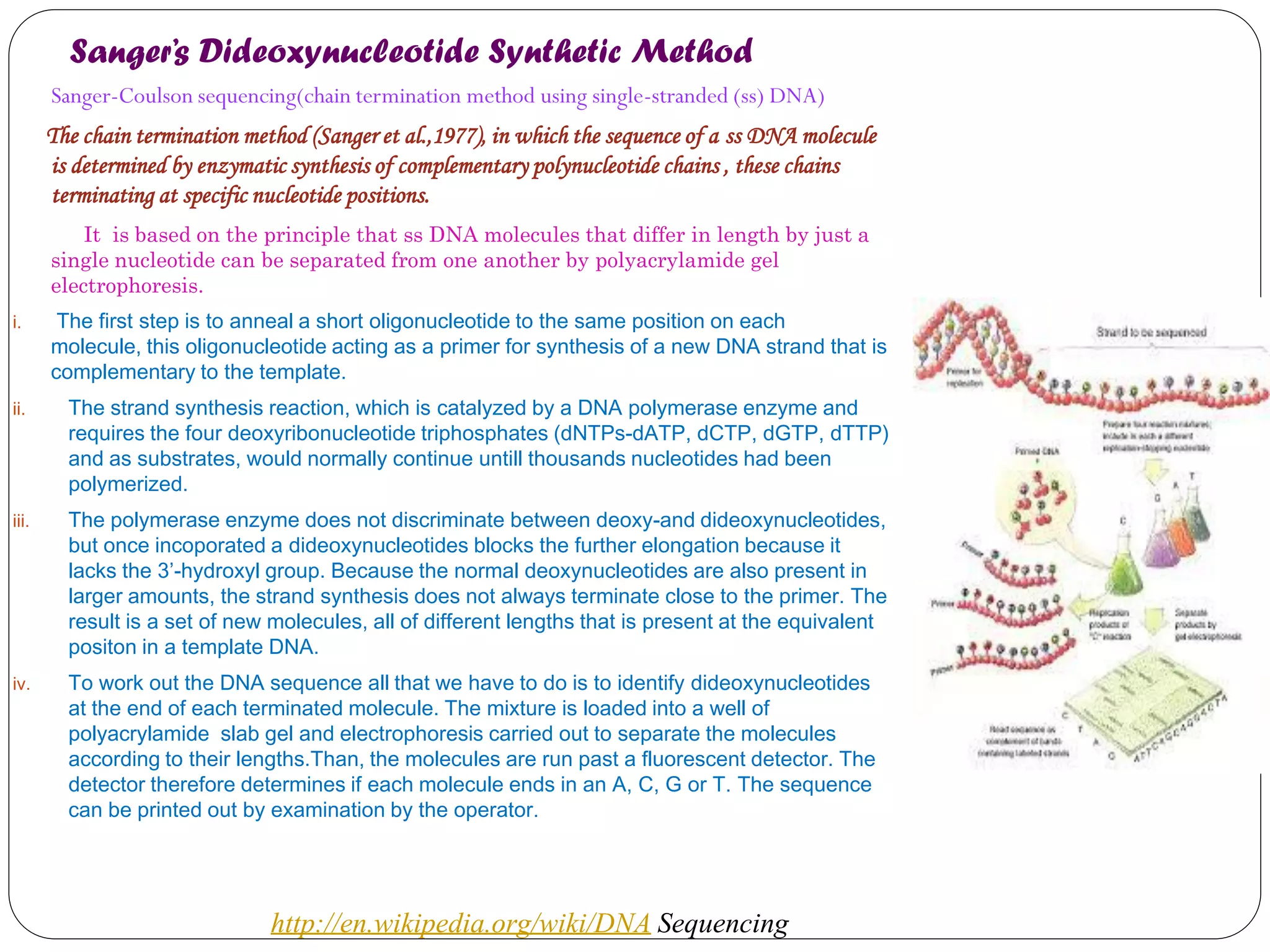
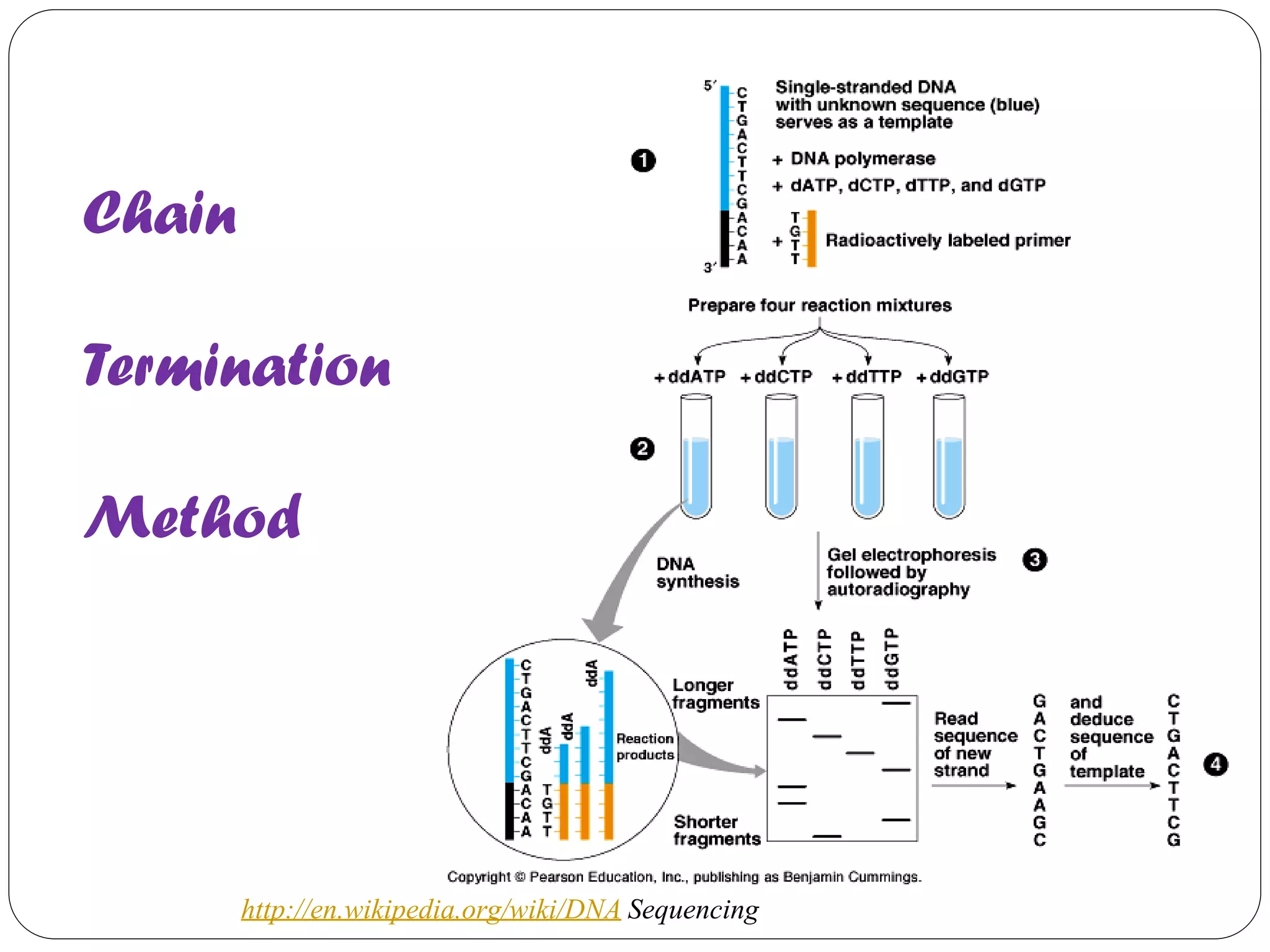
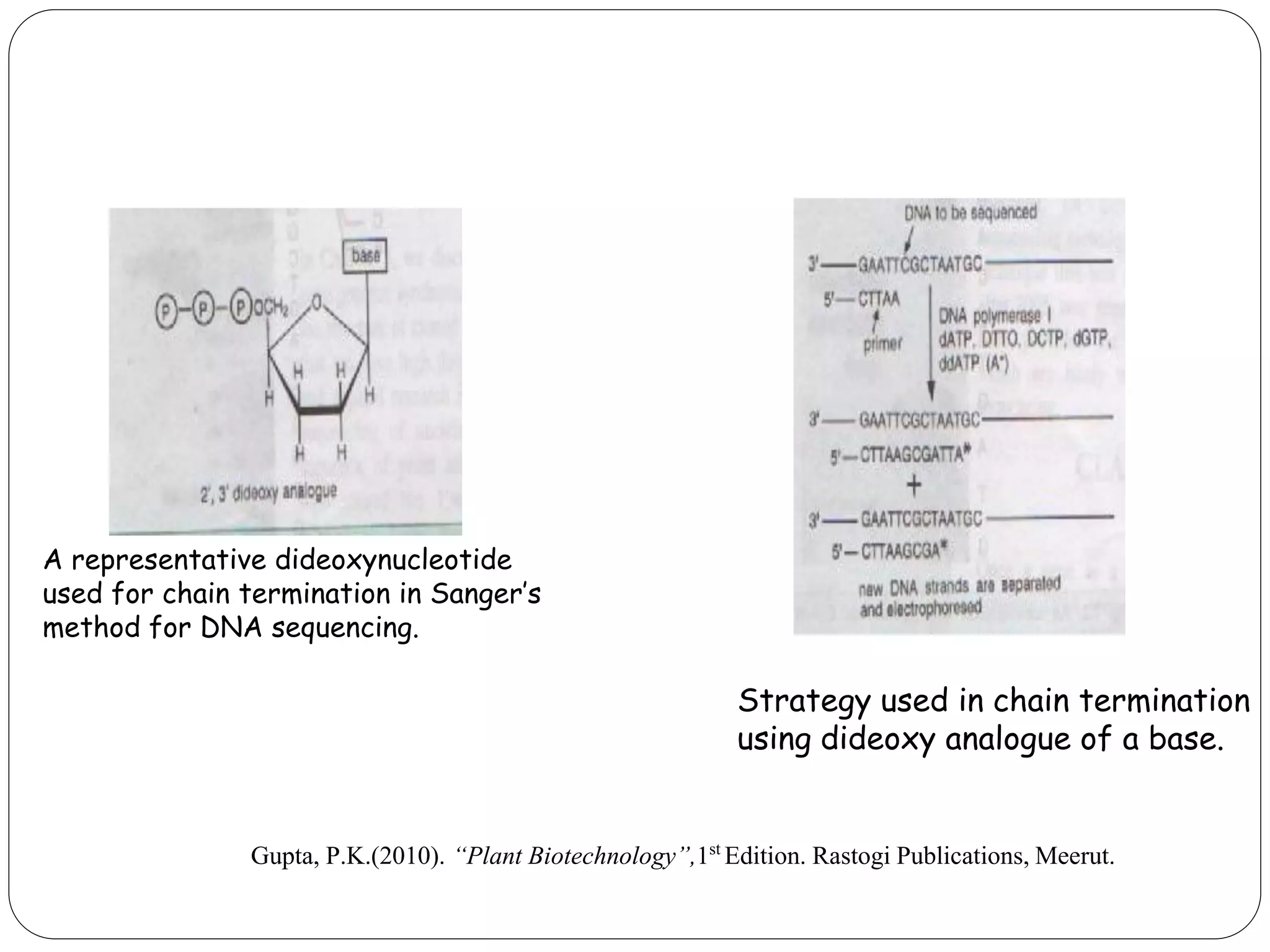
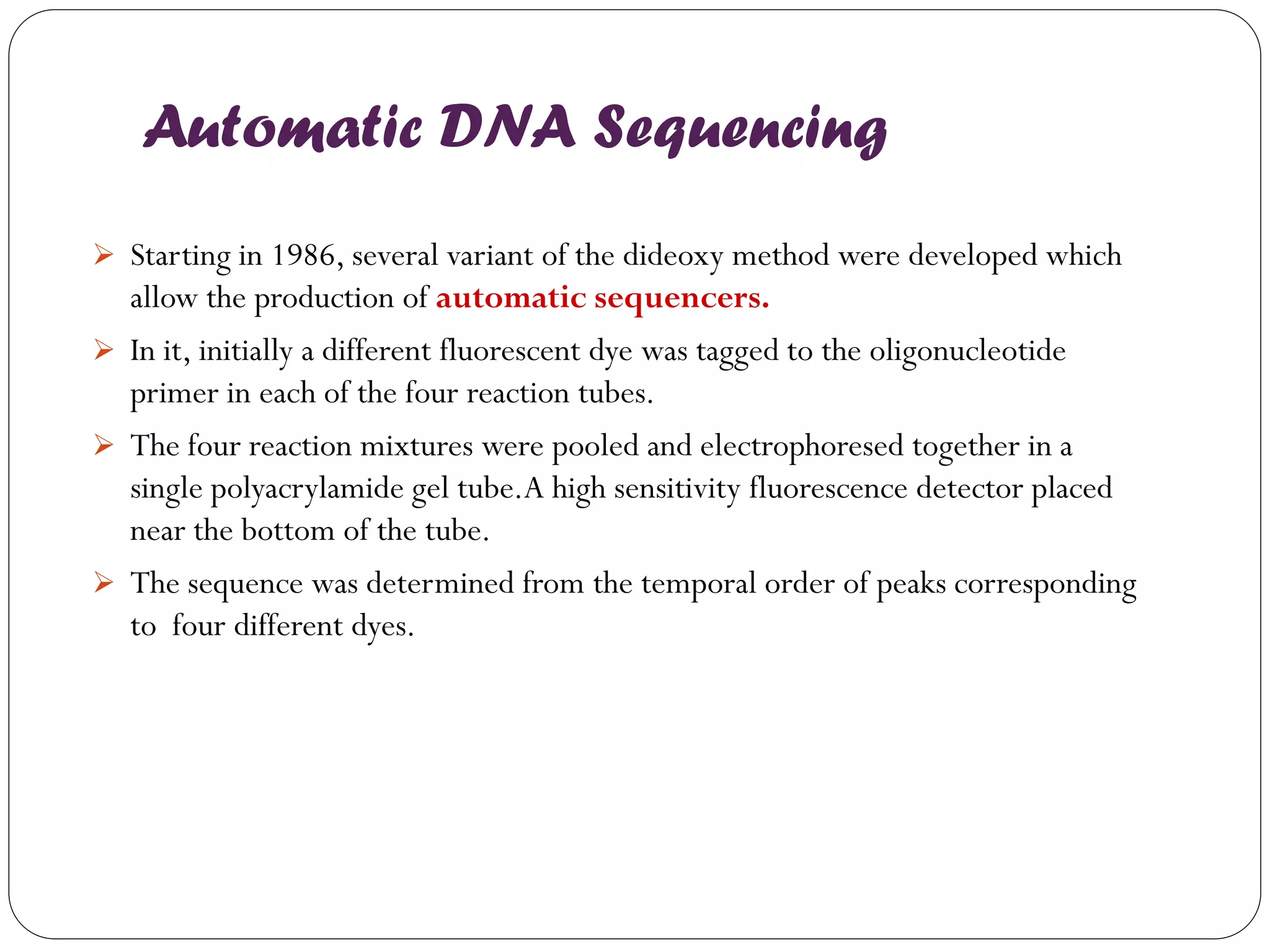
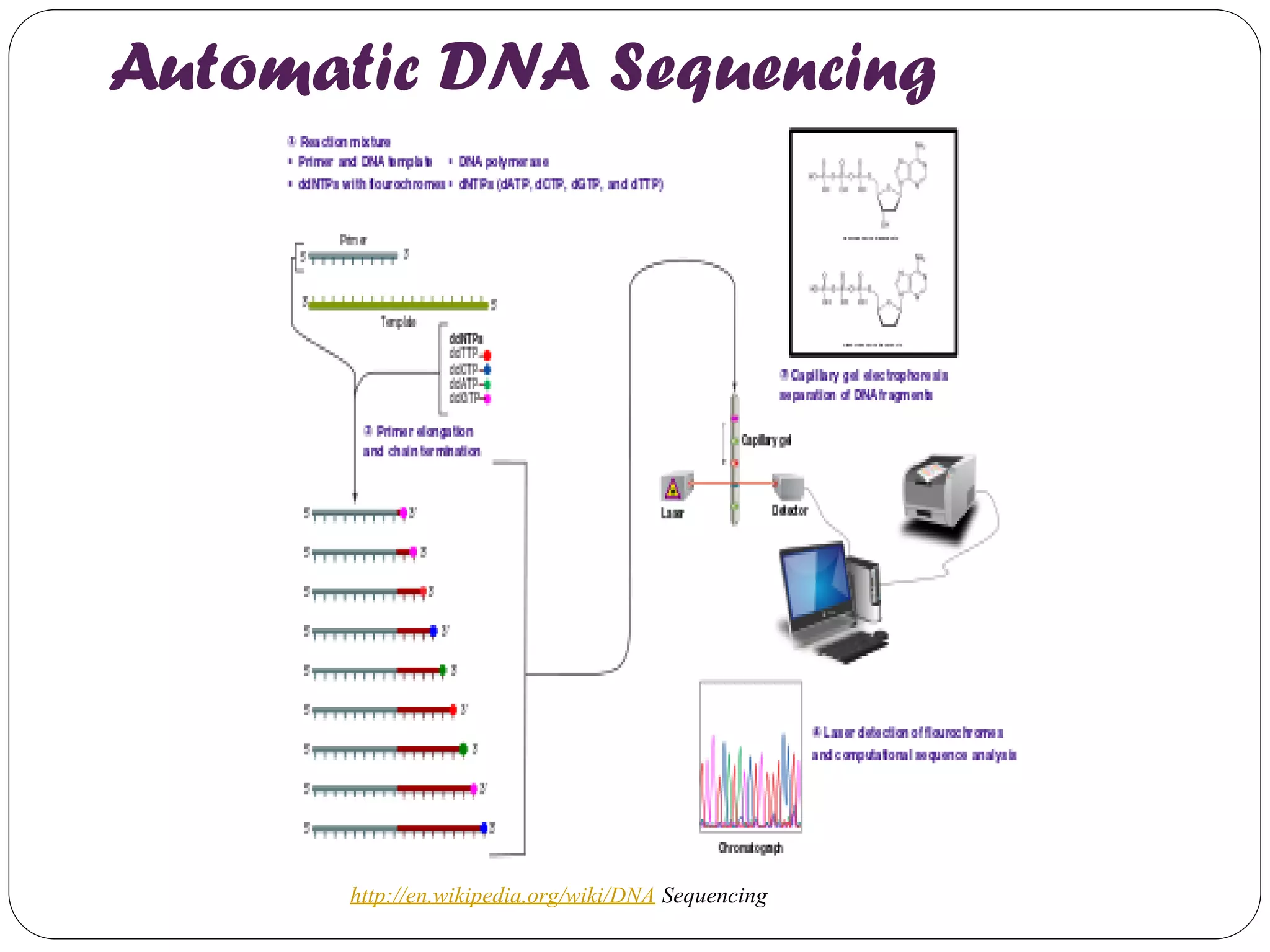
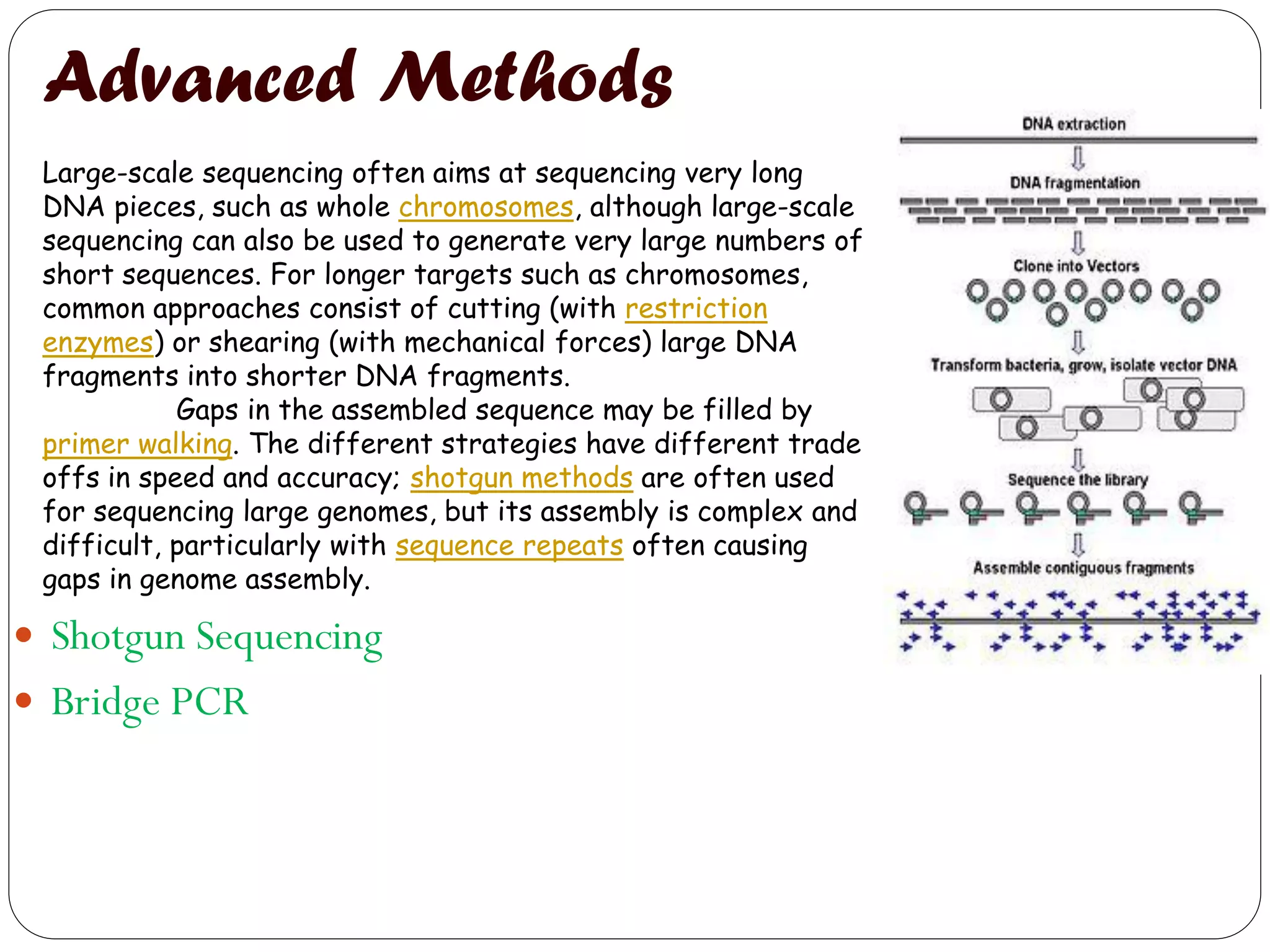
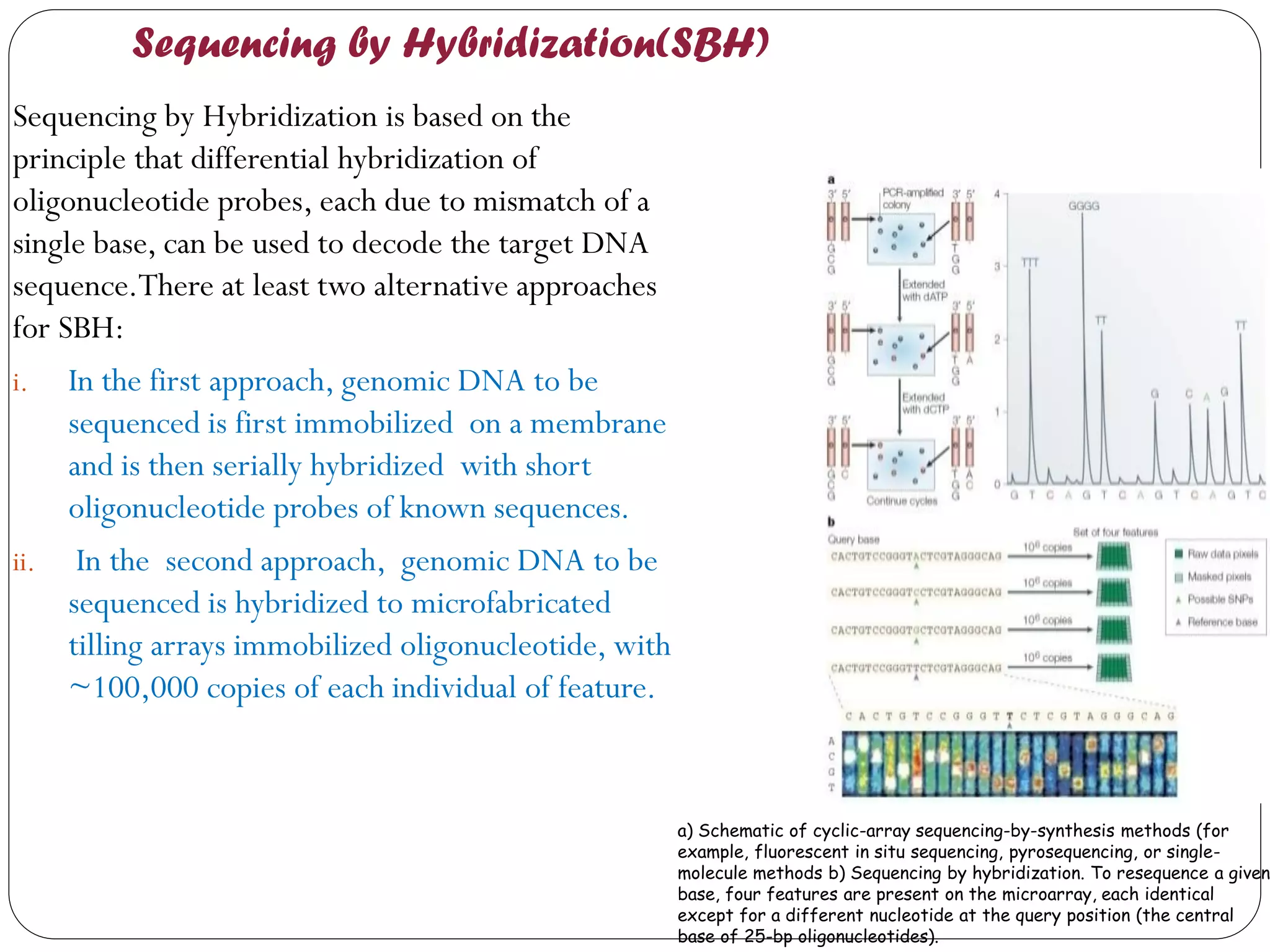
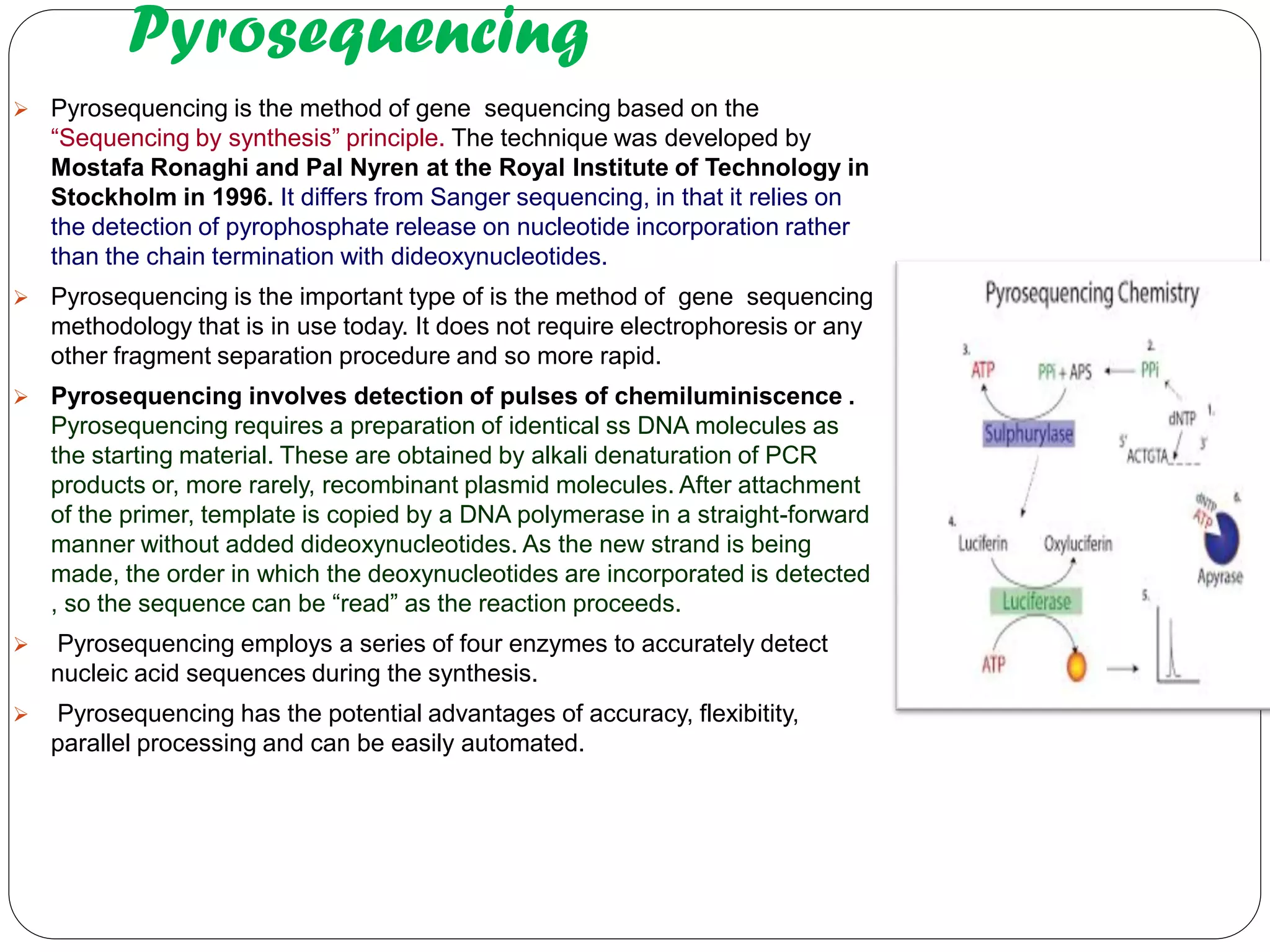
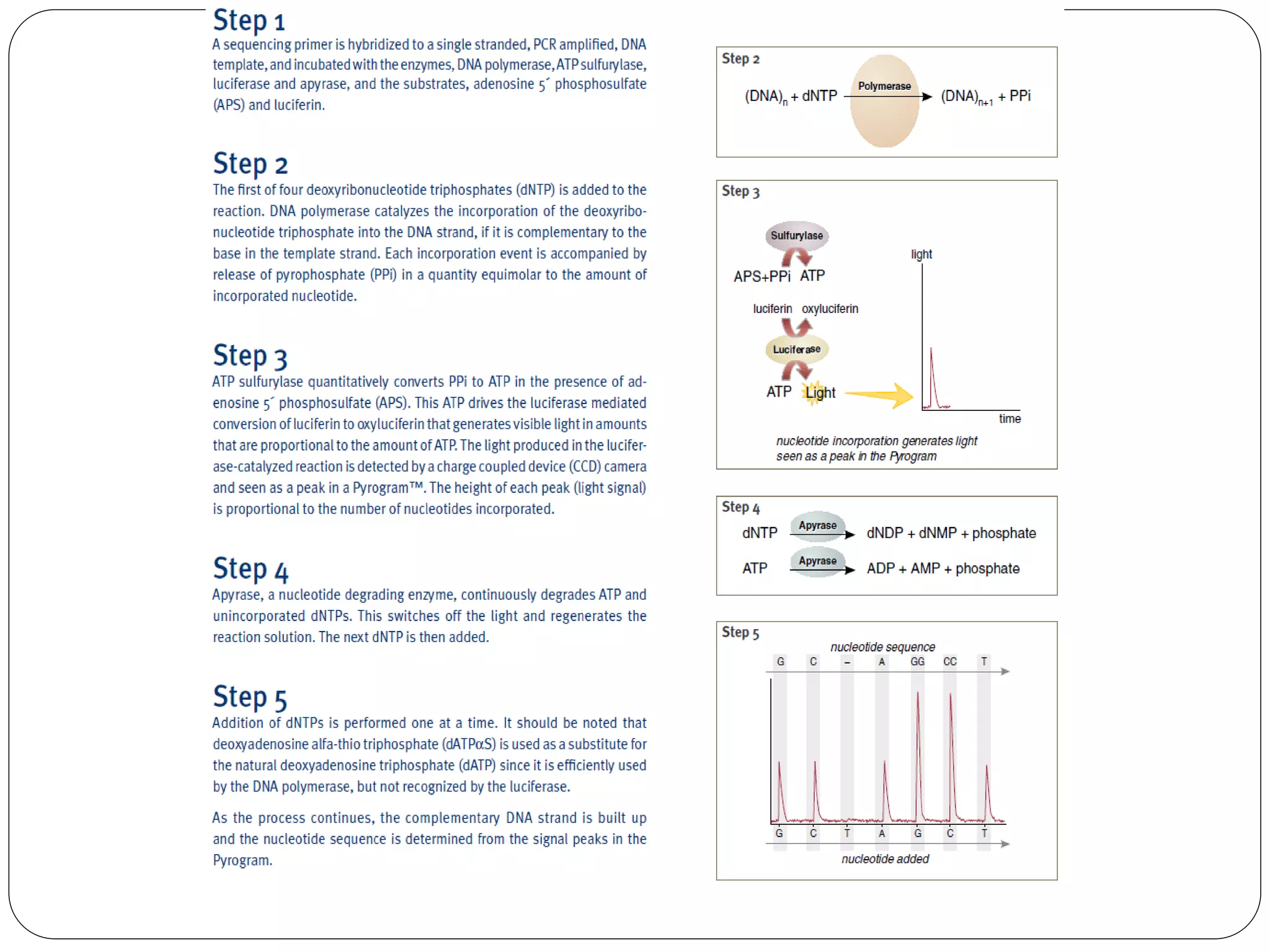
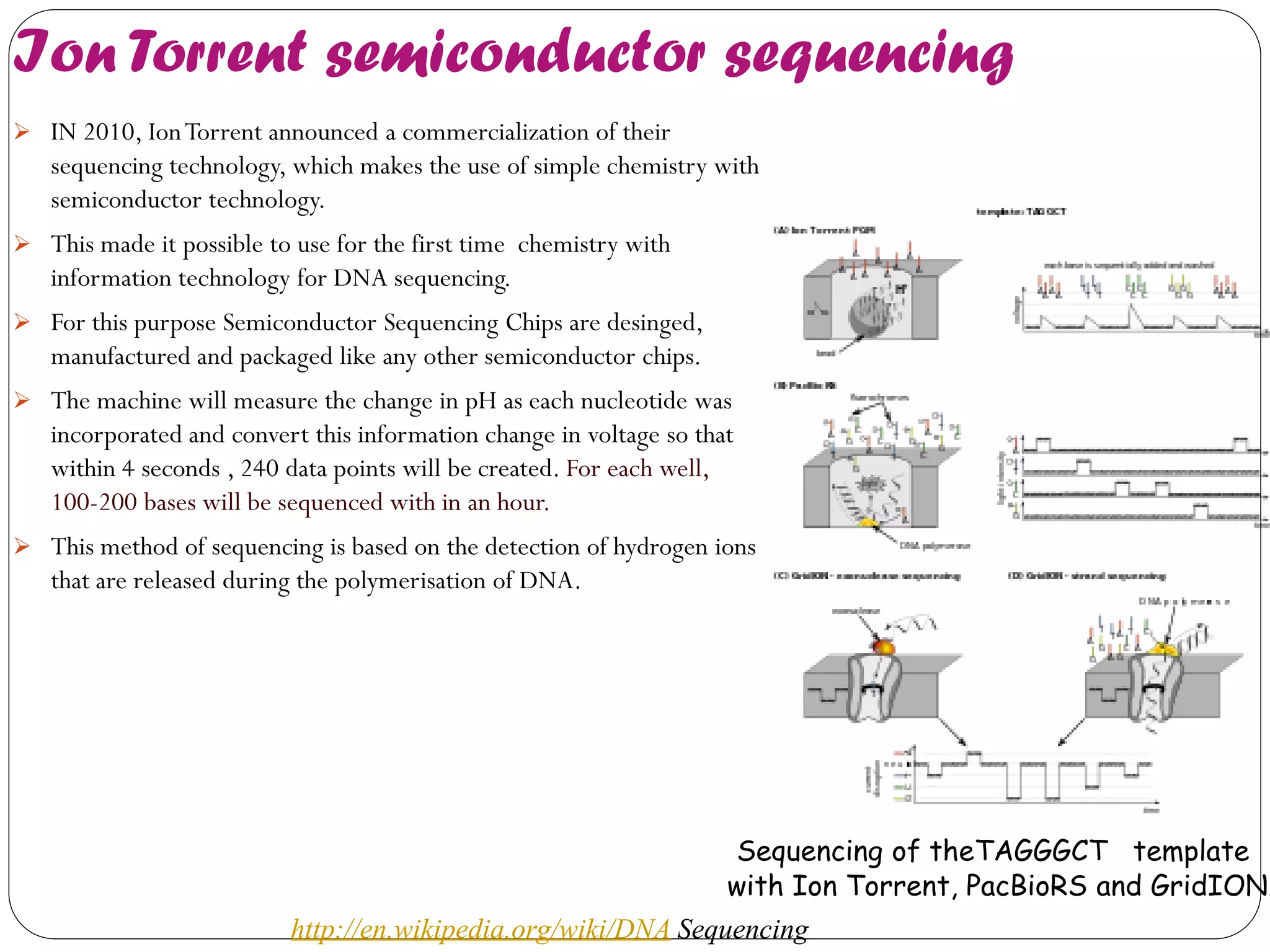
The document discusses various methods of gene sequencing. It begins with classical methods like Maxam-Gilbert chemical degradation and Sanger dideoxy chain termination. It then covers advanced methods such as shotgun sequencing and bridge PCR. The document also describes several new generation sequencing techniques, including sequencing by synthesis, pyrosequencing, nanopore sequencing, ion torrent sequencing, and Illumina sequencing. It provides details on the principles and procedures of many of these sequencing methods.