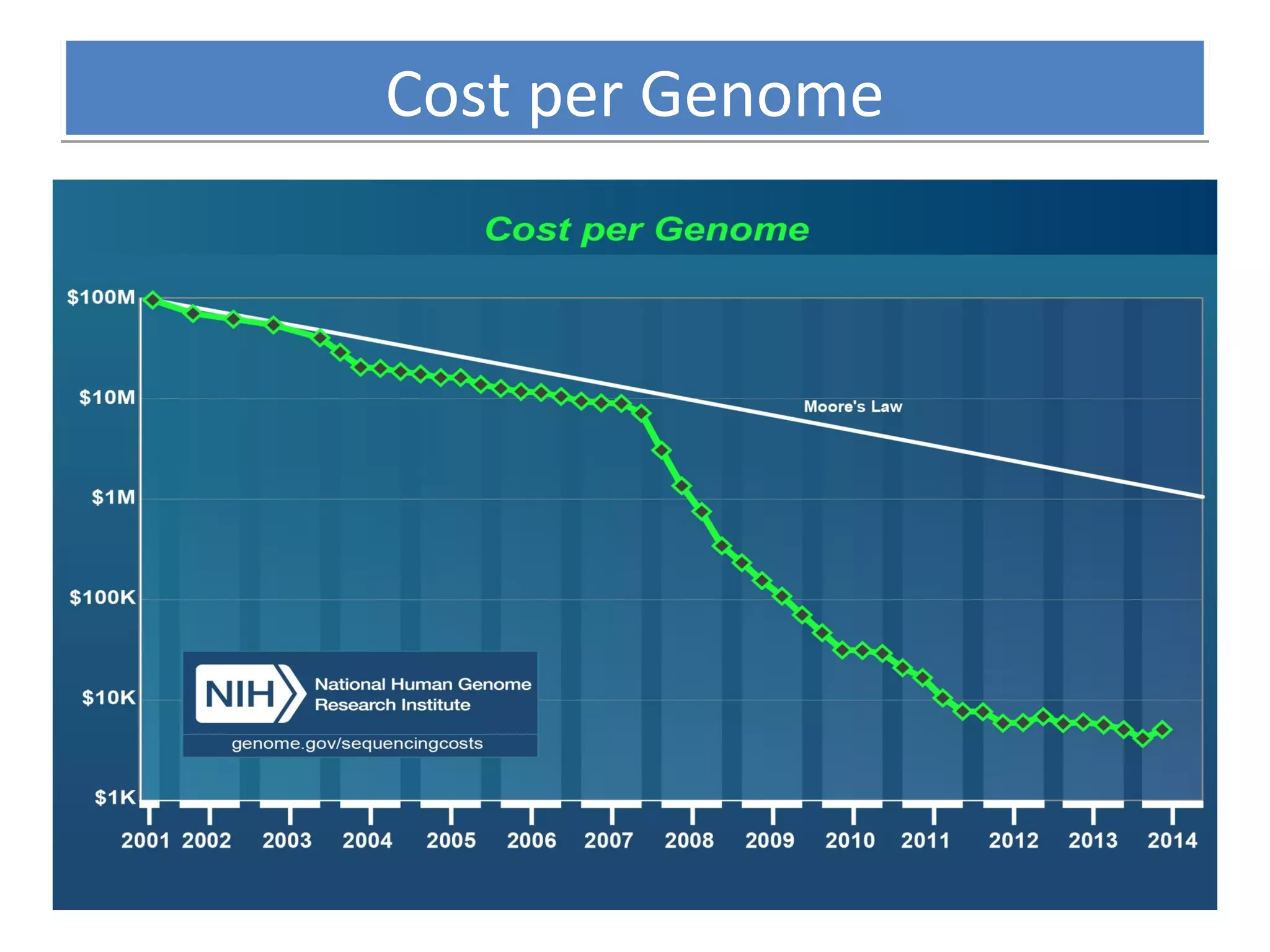
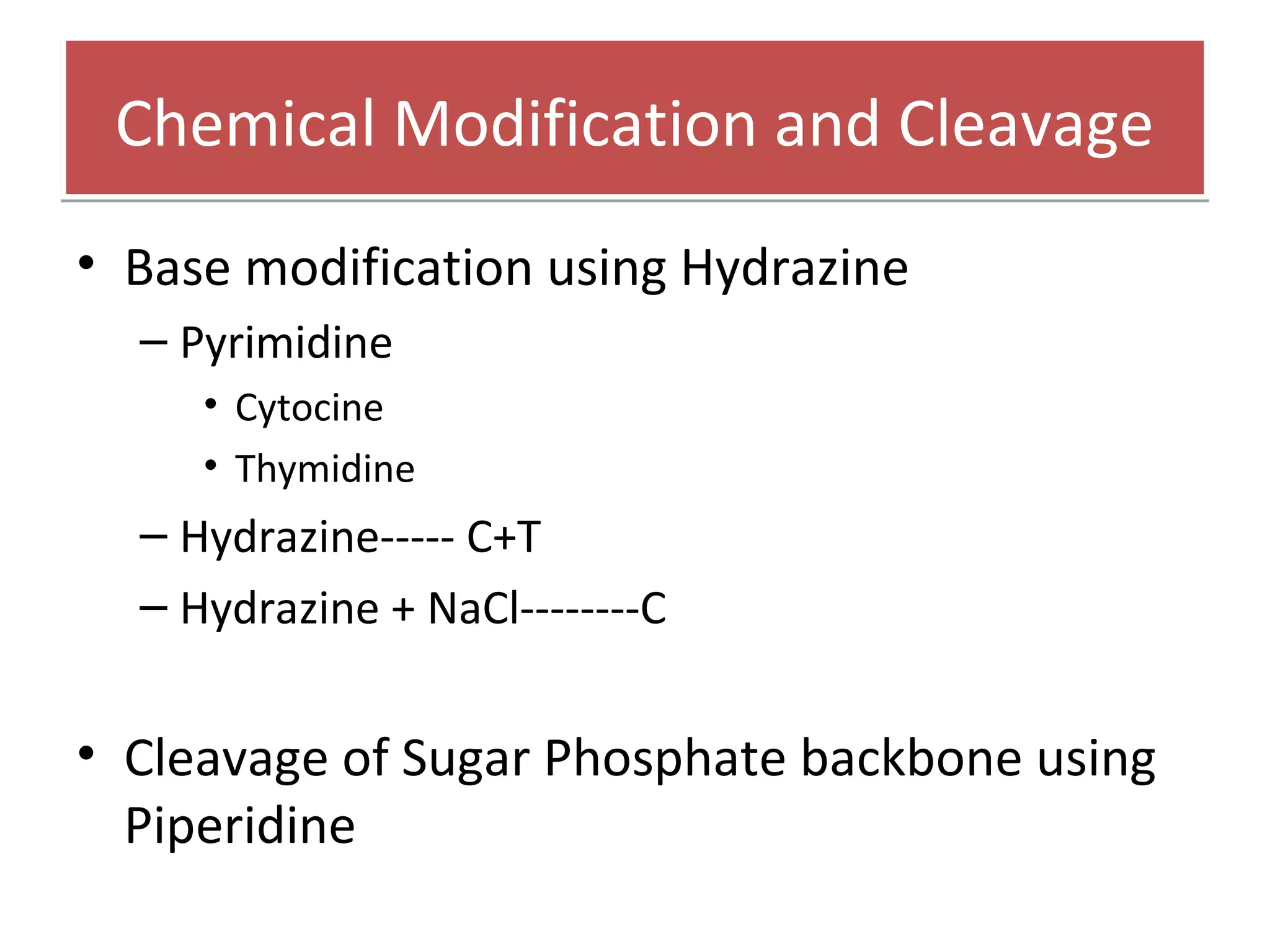
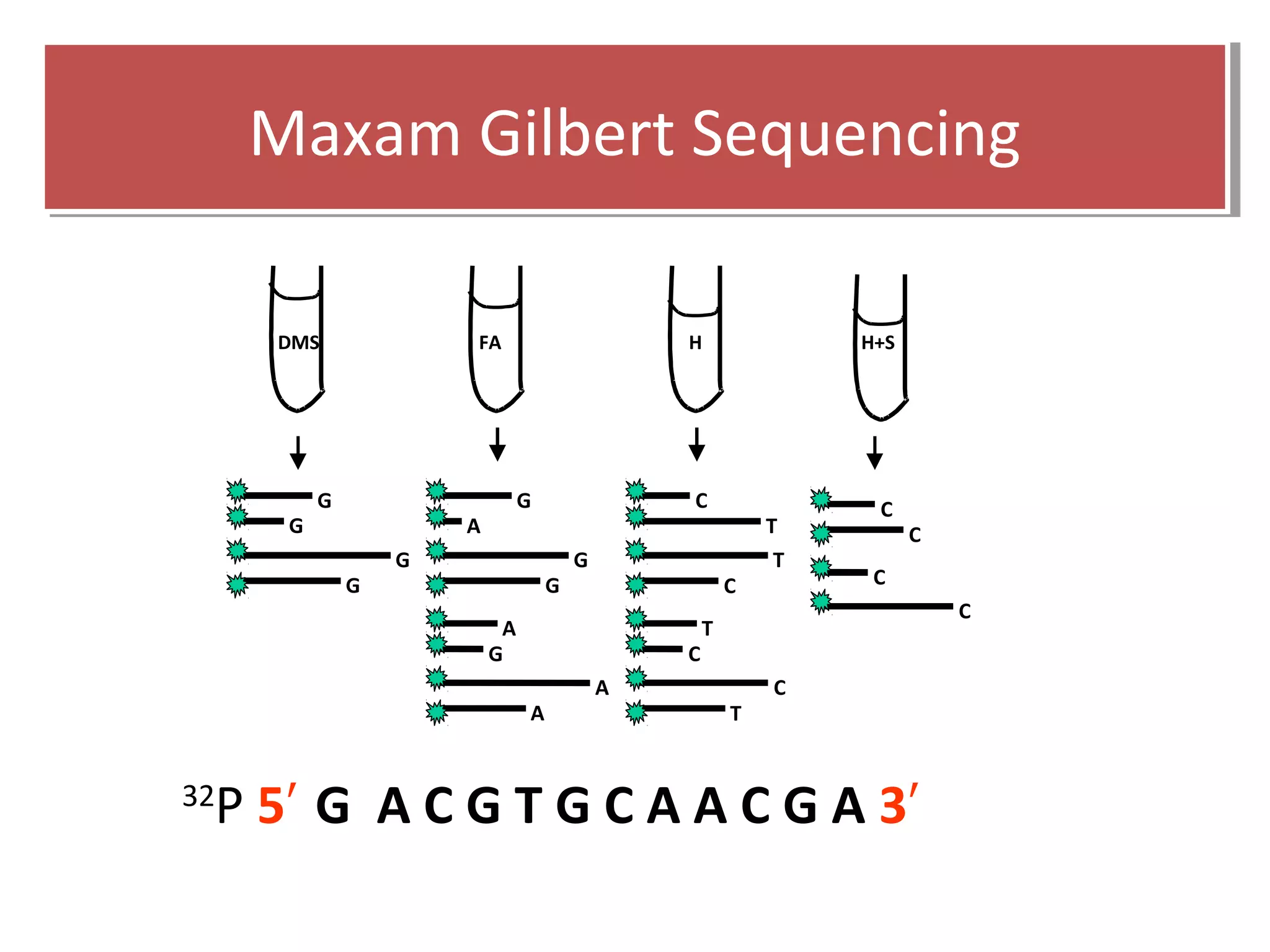
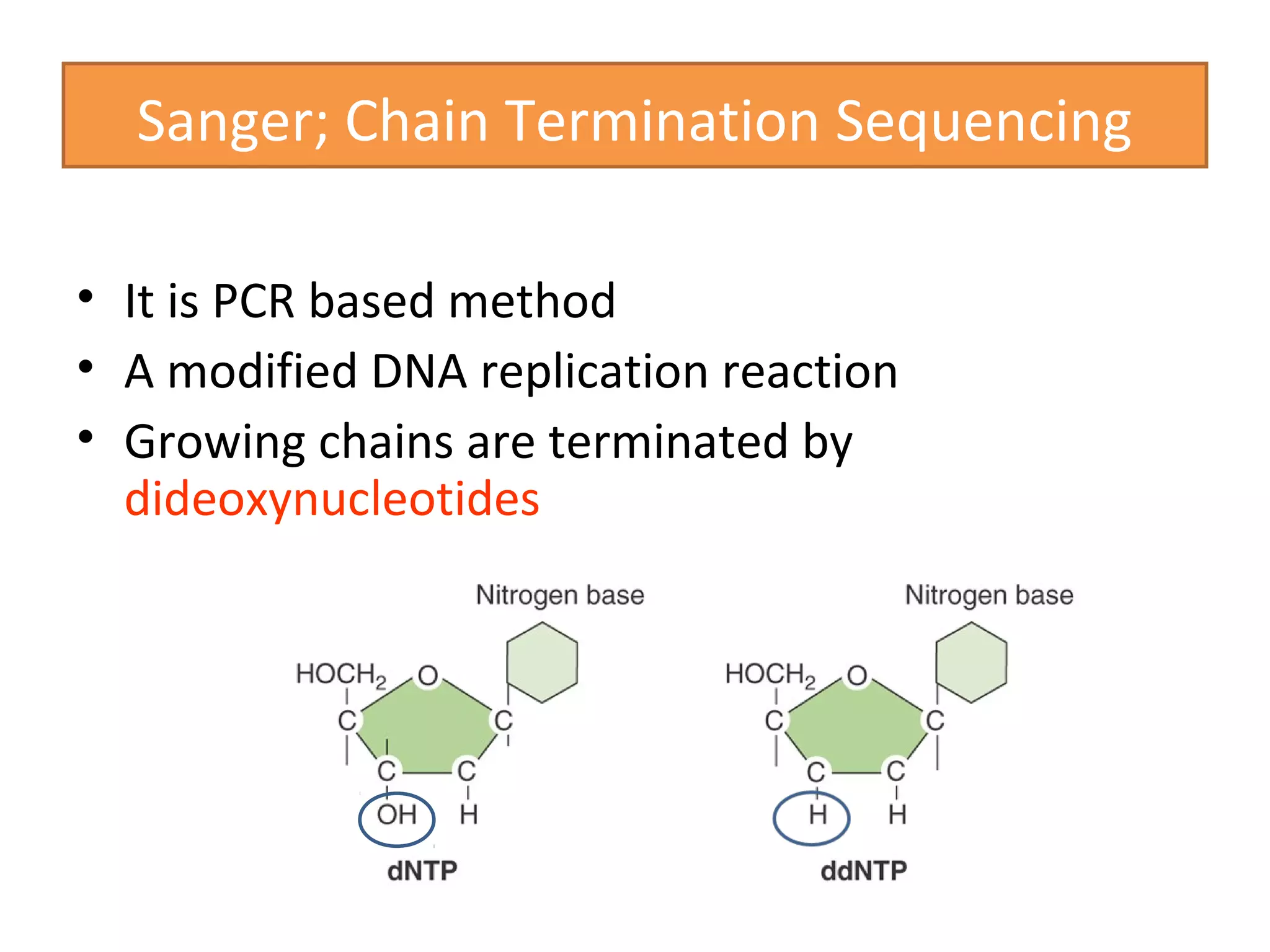
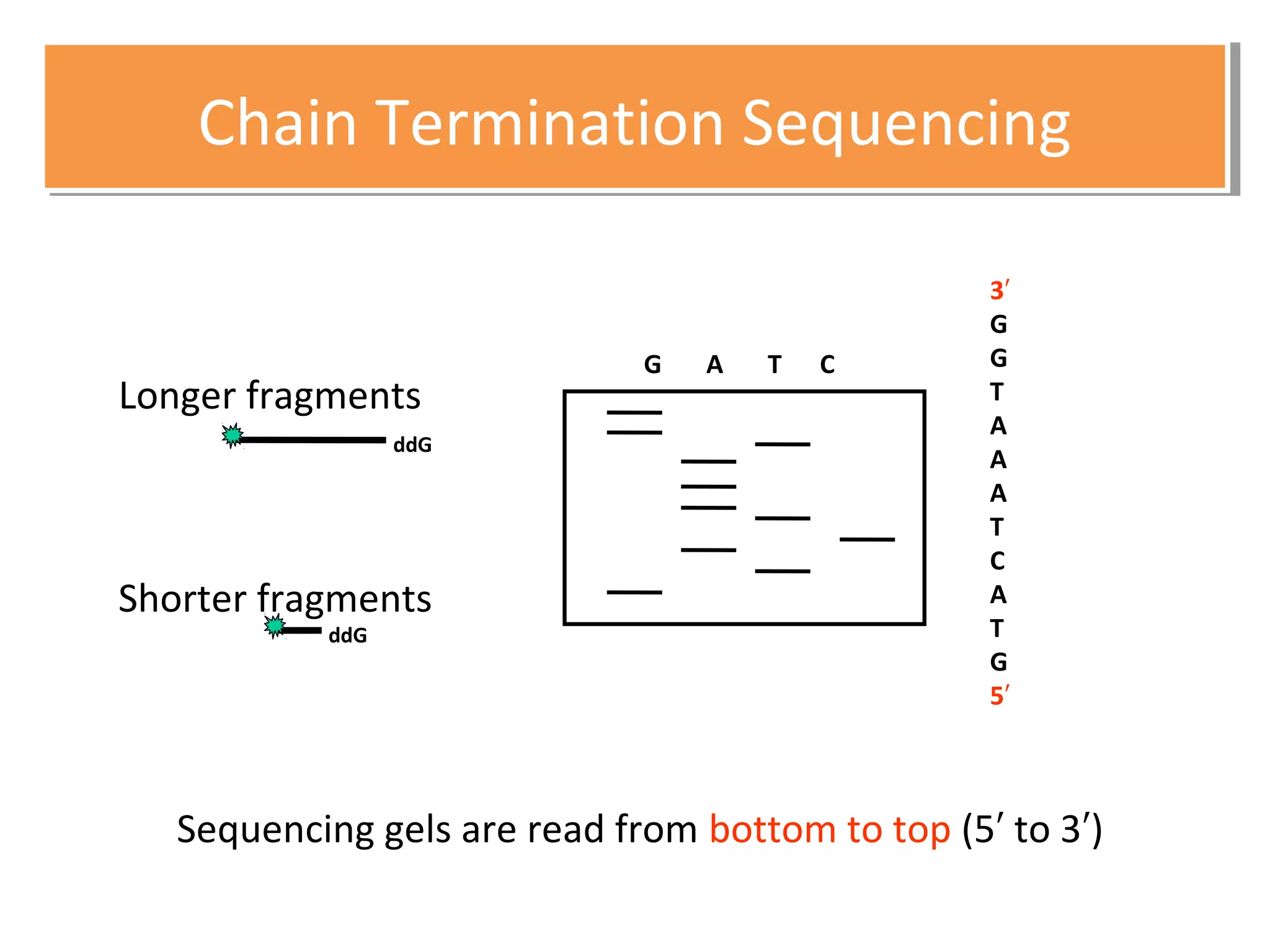
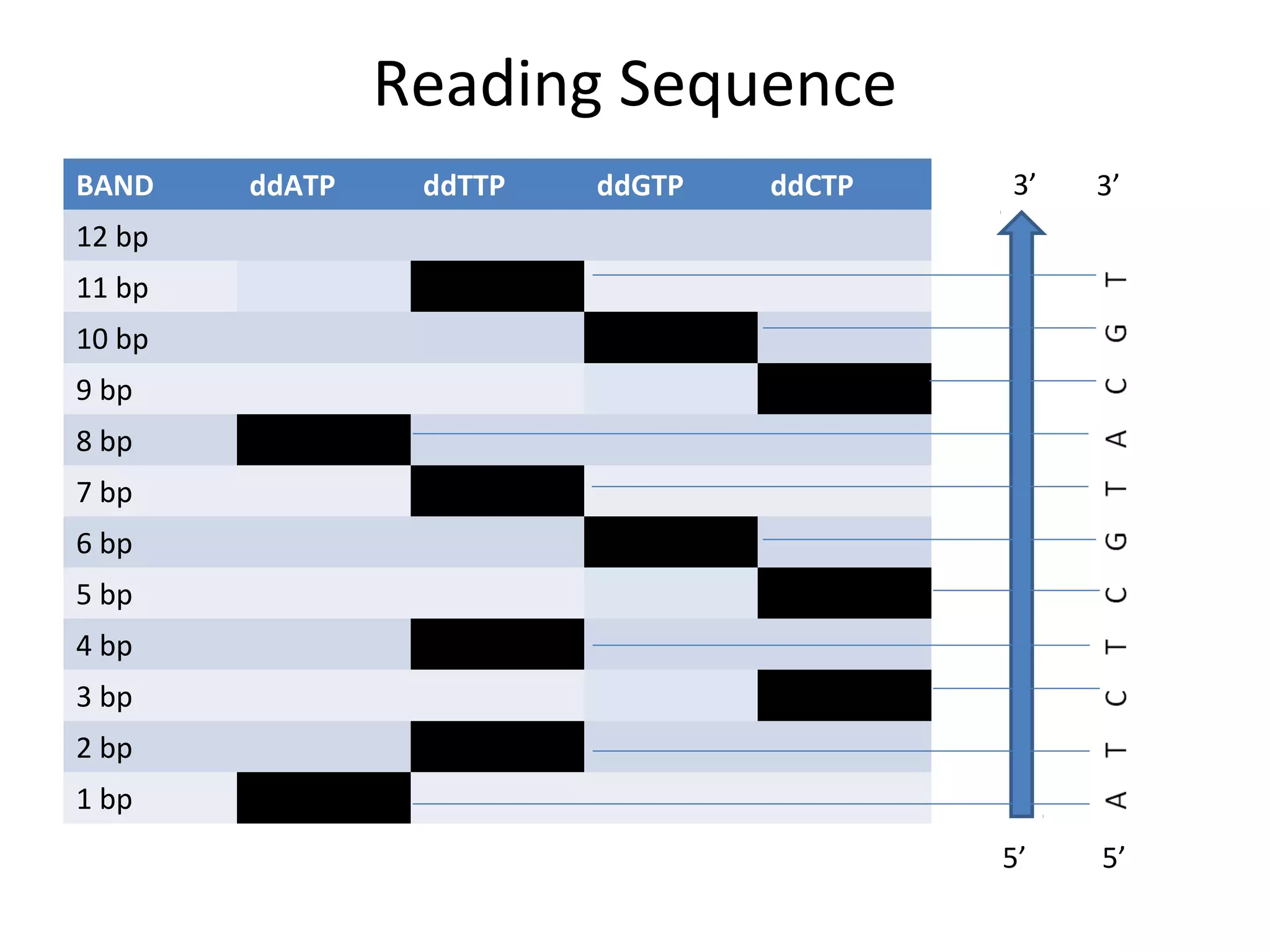
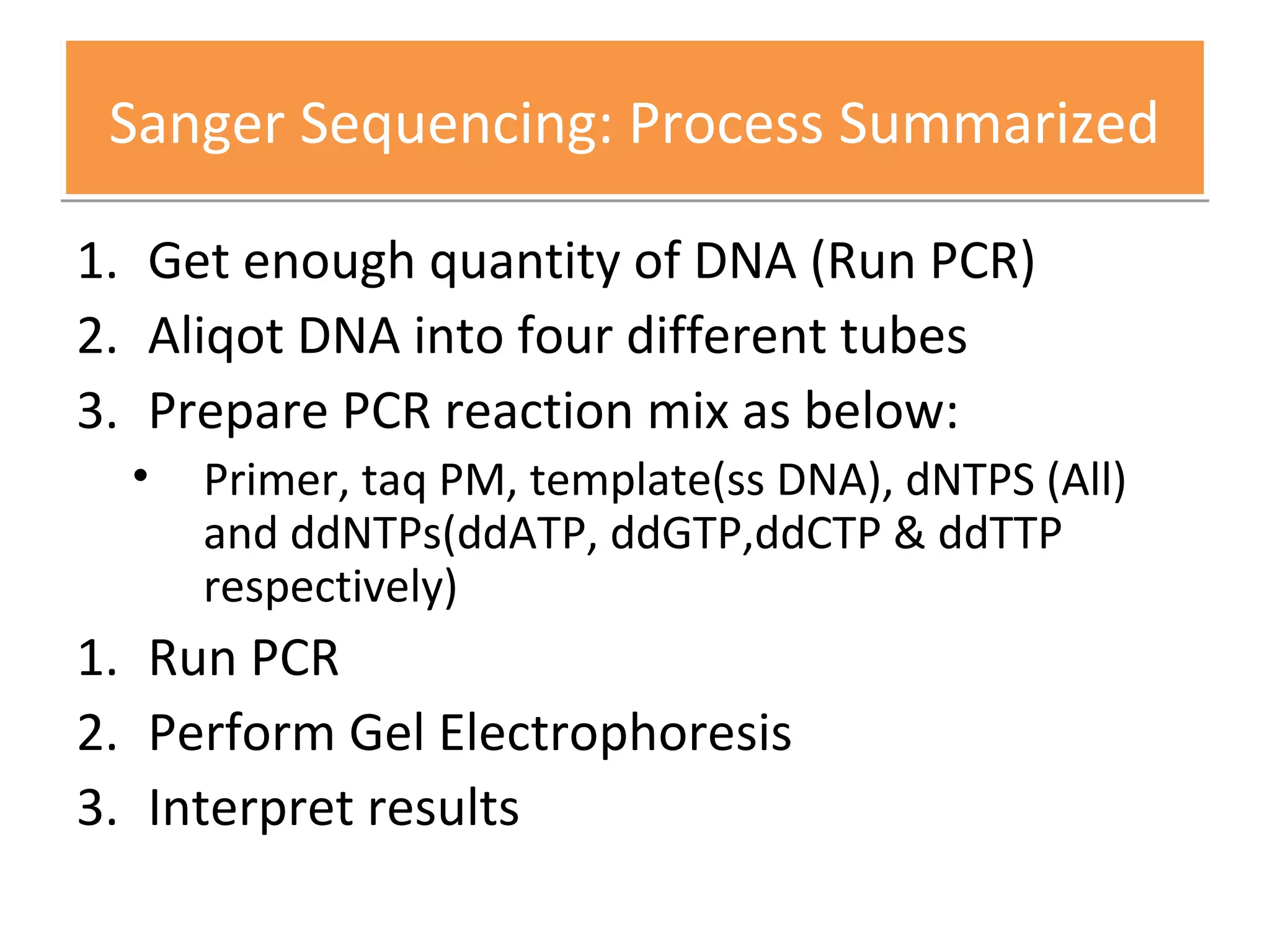
The document outlines DNA sequencing, highlighting its history, methods, and applications. It specifically details the chain termination method developed by Sanger and the chemical modification method established by Maxam and Gilbert. Additionally, it presents historical cost data trends for genome sequencing alongside a summary of each method's processes.