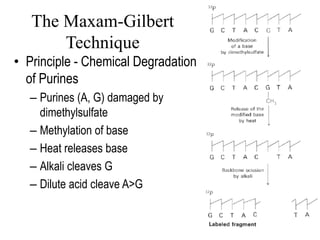
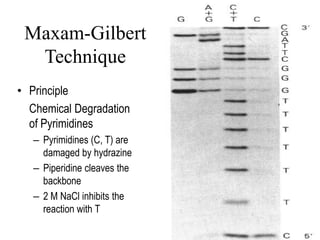
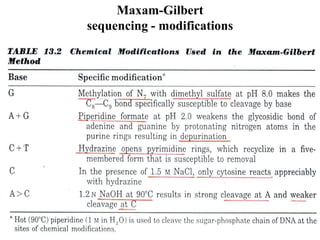
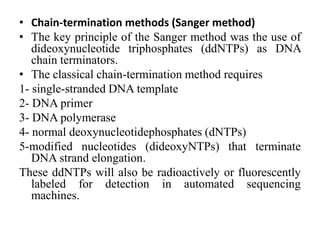
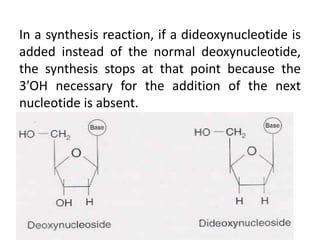
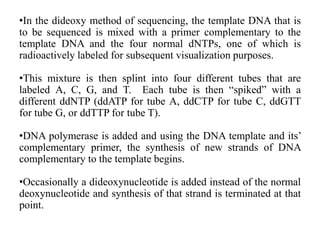
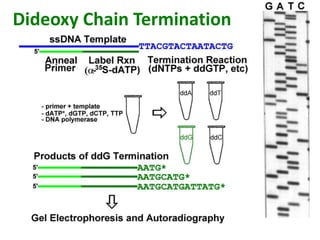
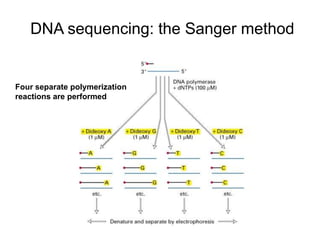
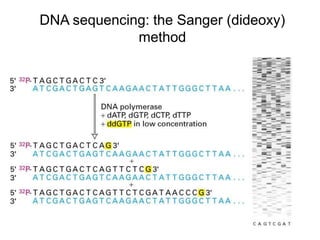
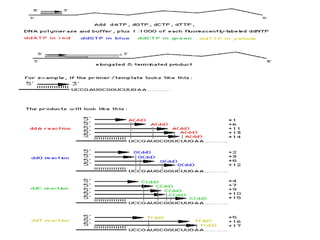
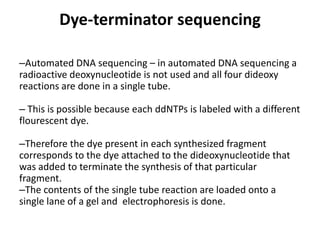
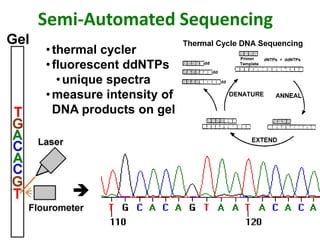
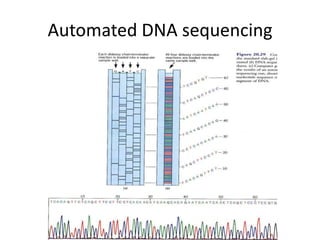
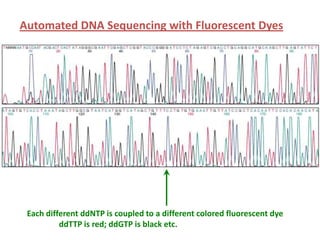
DNA sequencing involves determining the order of nucleotides in DNA. There are two main methods - the Maxam-Gilbert method uses chemical degradation to produce labeled DNA fragments of different lengths, while the Sanger method uses chain termination with dideoxynucleotides (ddNTPs) during DNA replication. The Sanger method involves 4 reactions with a normal nucleotide and one ddNTP each, producing fragments ending in each base. Electrophoresis separates the fragments by size, allowing the DNA sequence to be read from gel images. Modern automated sequencing uses fluorescently-labeled ddNTPs in a single reaction tube analyzed by a fluorimeter.