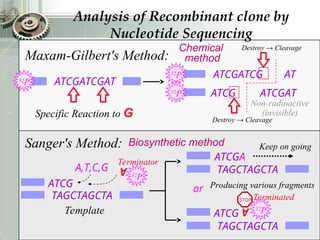
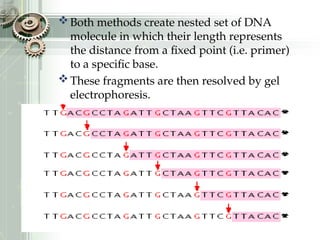
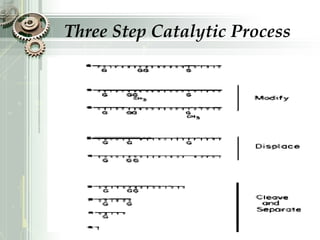
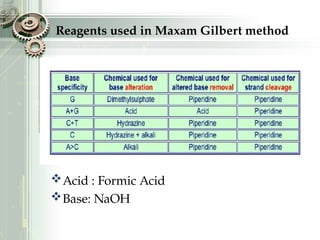
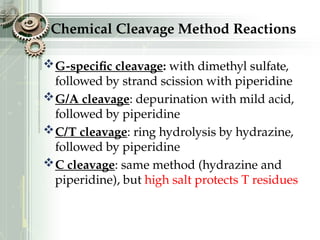
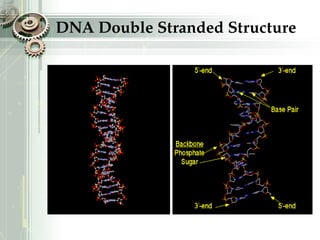
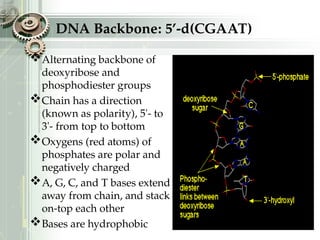
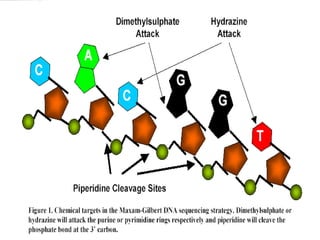
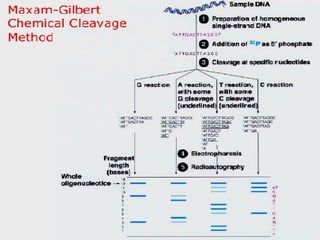
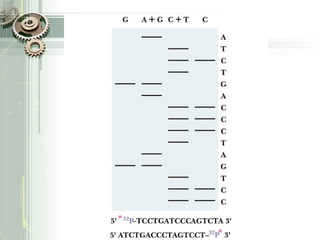
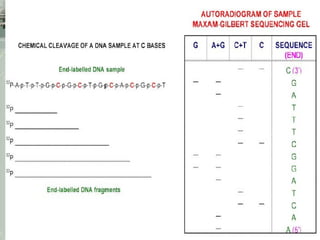
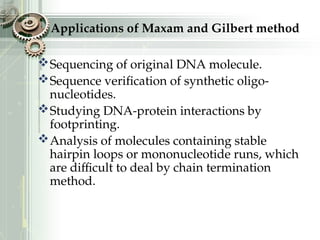
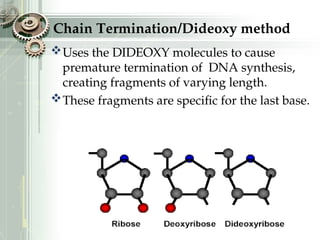
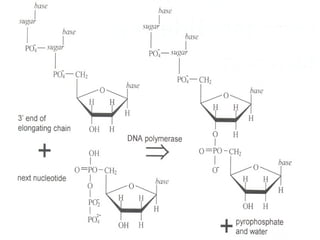
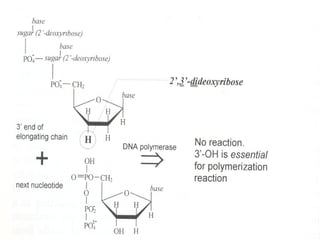
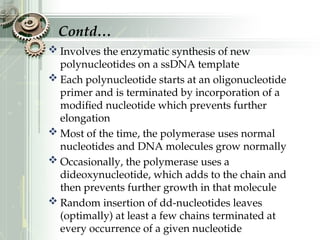
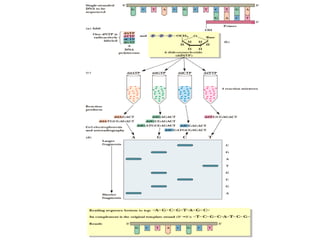
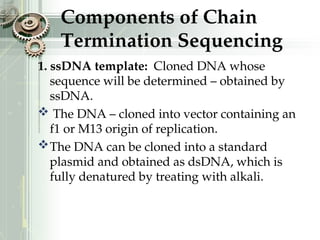
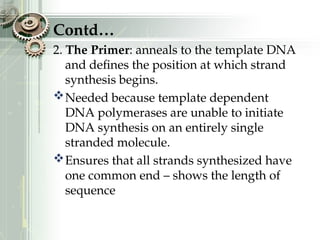
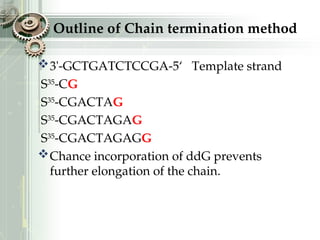
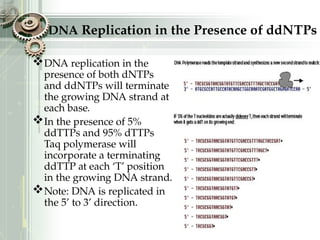
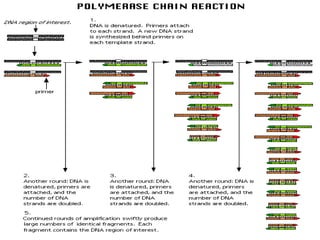
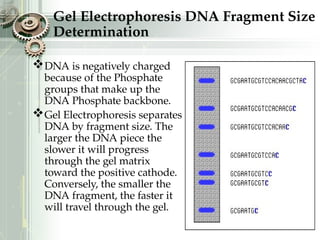
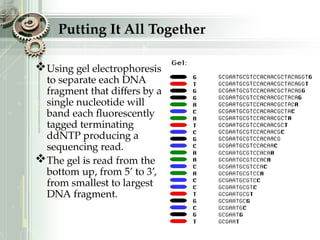
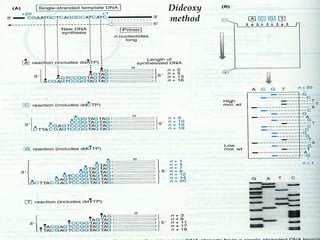
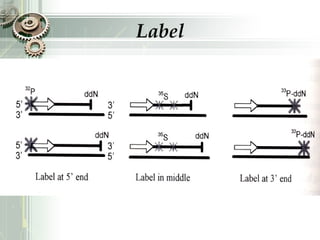
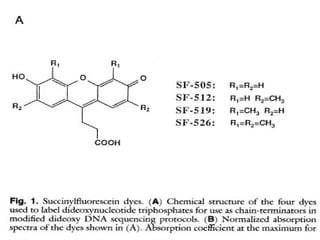
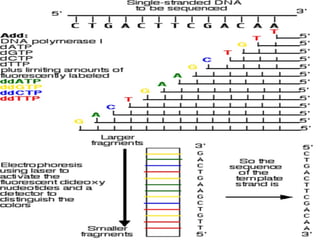
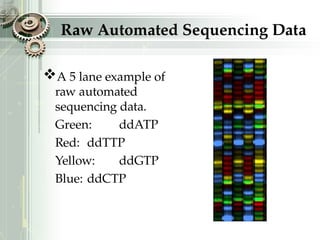
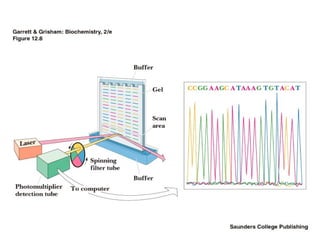
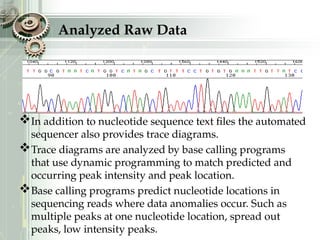
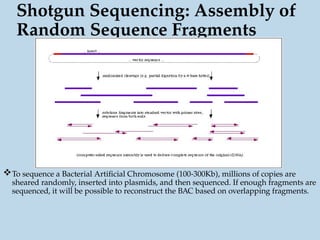
DNA sequencing is the process of determining the nucleotide sequence of a DNA molecule, which helps in understanding its structure and function. Two primary methods used for DNA sequencing are the Maxam-Gilbert chemical degradation method and the Sanger chain termination method, each having unique advantages and disadvantages. Recent advancements include automated sequencing techniques with fluorescent labeling, enabling efficient analysis of DNA fragments through gel electrophoresis.