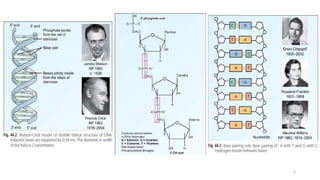
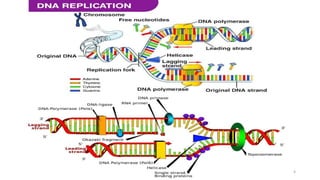
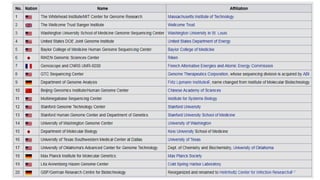
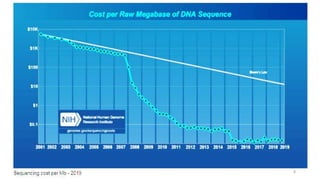
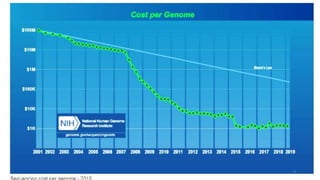
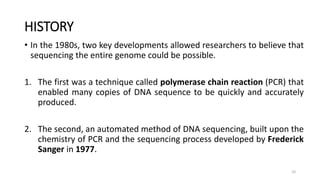
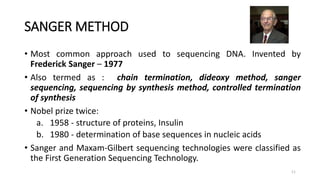
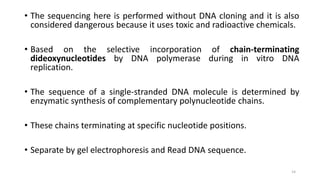
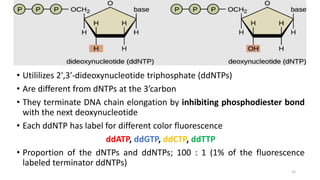
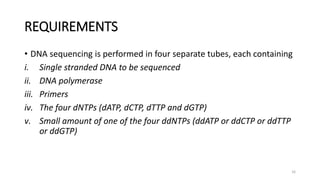
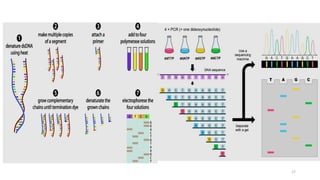
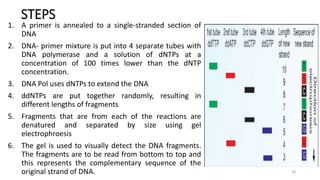
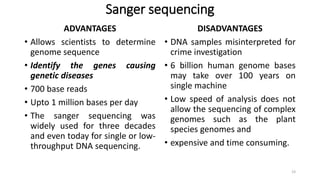
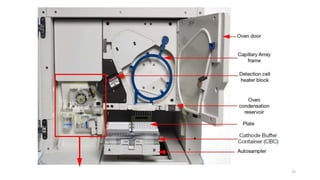
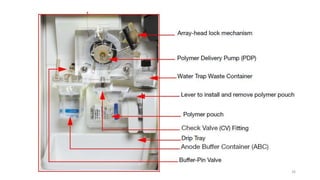
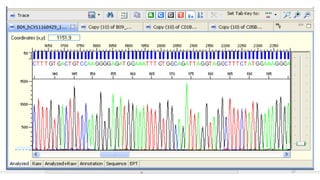
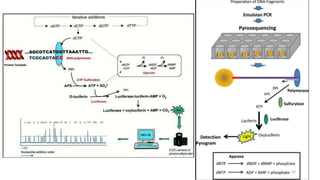
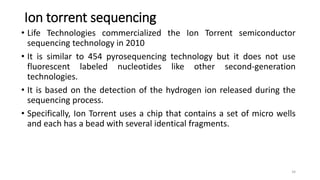
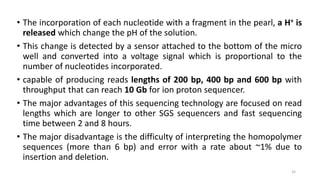
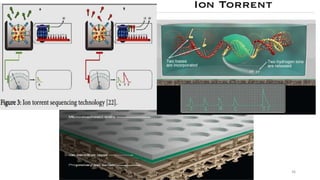
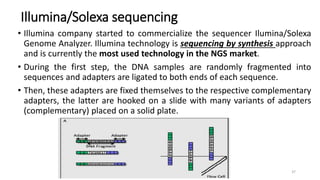
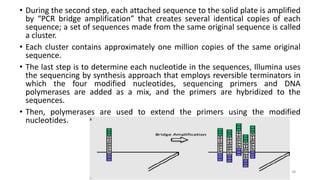
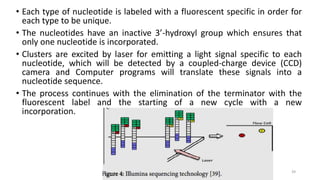
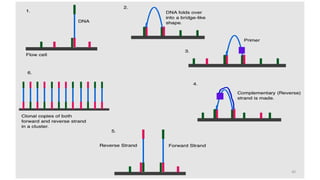
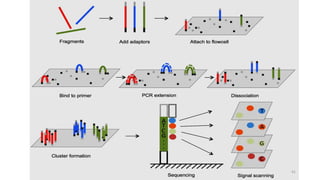
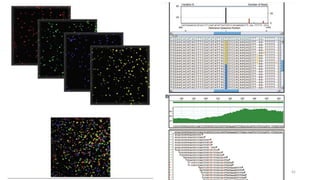
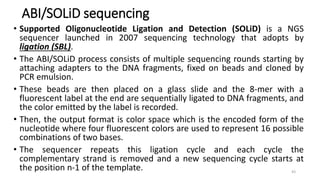
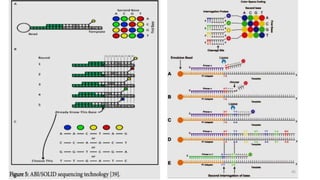
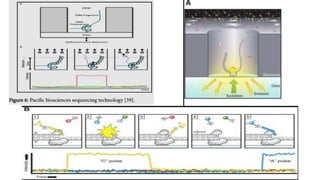
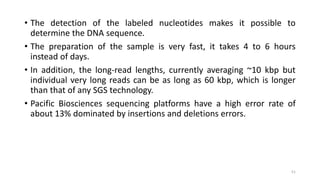
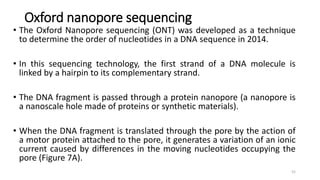
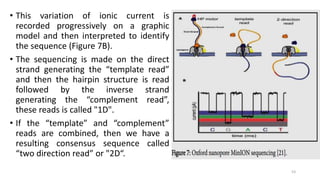
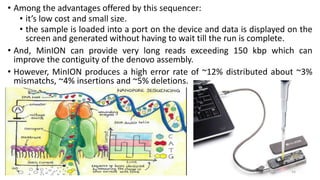
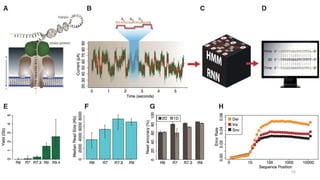
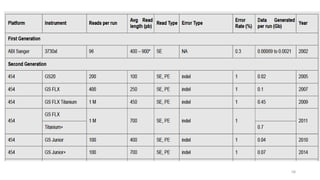
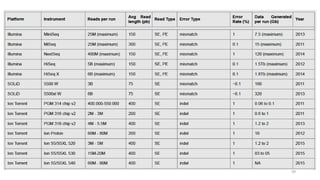
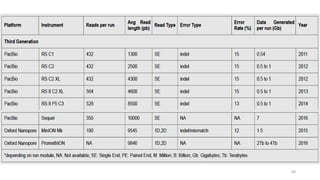
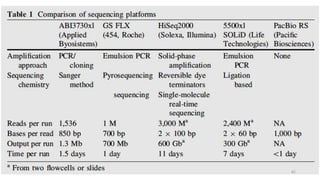
The document provides a comprehensive overview of DNA sequencing, detailing its history, methodologies, and advancements including the Human Genome Project and various sequencing technologies such as Sanger, Maxam-Gilbert, and second and third generation techniques. It outlines the benefits and limitations of these technologies, emphasizing the evolution from manual methods to automated processes that enhance speed and reduce costs. The content serves as a technical guide for understanding DNA sequencing's impact on genetics and the biological sciences.