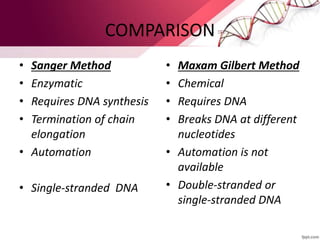
The document discusses Sanger sequencing, a method of DNA sequencing. It provides a brief history of DNA sequencing, noting that Sanger developed an enzymatic DNA sequencing technique in 1977. The document then describes the key steps of Sanger sequencing, including separating the DNA strands, copying one strand with chemically altered bases that cause termination, and analyzing the fragments to reveal the DNA sequence. It also compares Sanger sequencing to the Maxam-Gilbert chemical degradation method.