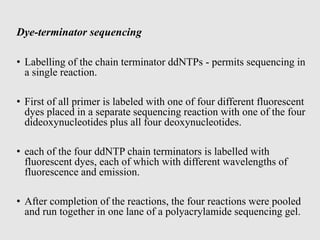
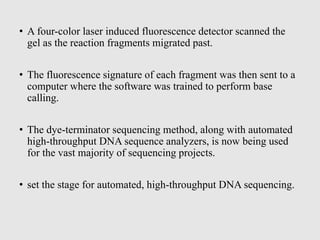
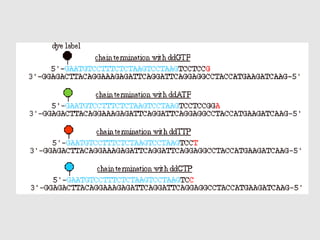
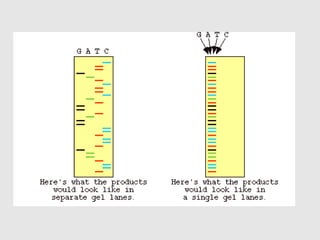
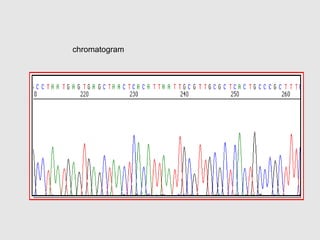
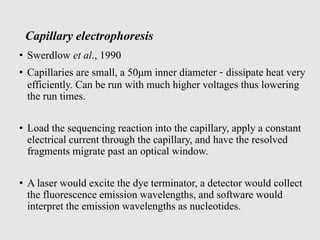
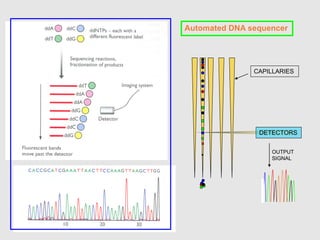
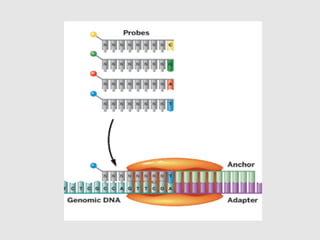
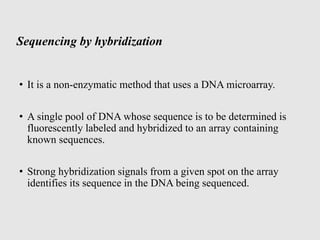
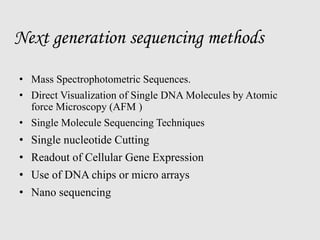
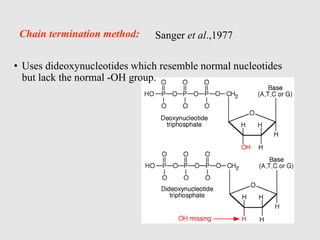
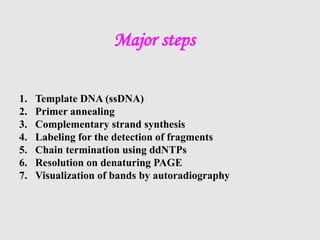
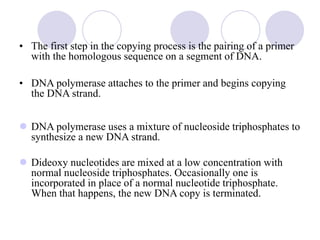
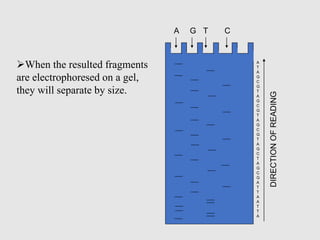
This document discusses various gene sequencing methods. It begins by introducing DNA and the importance of sequencing the genetic code. It then describes several early sequencing techniques like Sanger sequencing using chain termination or chemical cleavage. It discusses the need for sequencing to understand genetic conditions. The document also covers topics like genome sequencing, genomics, and high-throughput sequencing techniques like dye-terminator sequencing which replaced radioactive labels with fluorescent labels to automate the process.













![Cycle sequencing
Thus, less template DNA is needed than for conventional
sequencing reactions.
Furthermore, the repeated heating and cooling can be done in a
DNA thermal cycler.
Advantages:
•Works with ssDNA and dsDNA and thus eliminates the need for
M13 phage
•Requires only small amounts of template
•Can be set up in microtitre plates or microtubes
•Can use internal labeling with [α-32P], [α-33P],or [35S]or with 5’-
end labeled primer
•Can be adapted for rapid screening](https://image.slidesharecdn.com/genesequencingmethods-180430094002/85/Gene-sequencing-methods-24-320.jpg)