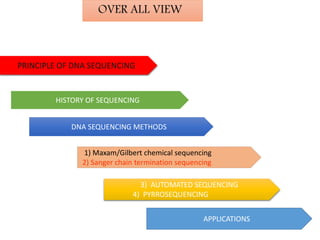
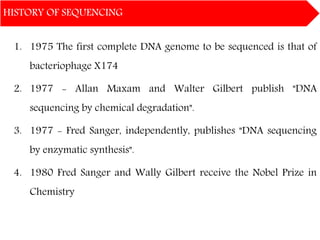
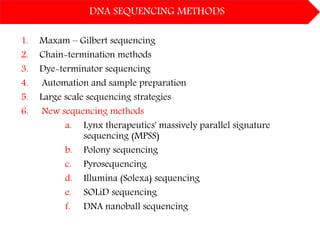
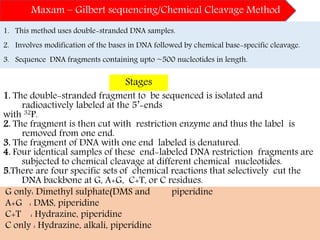
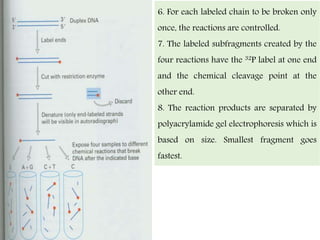
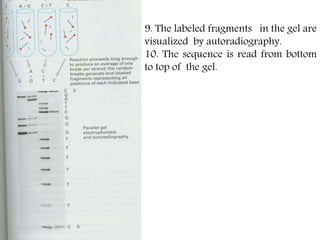
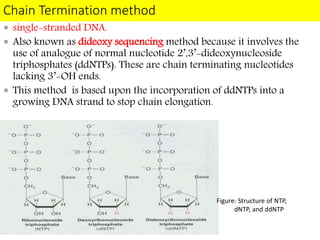
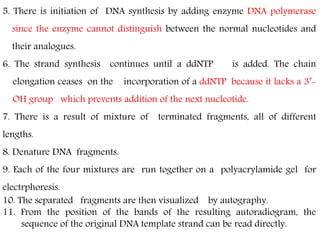
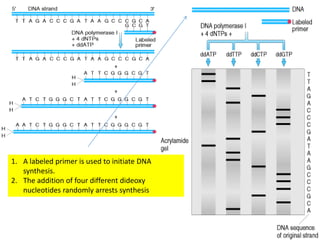
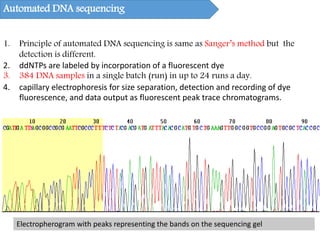
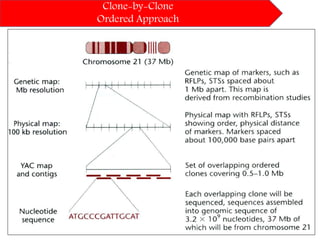
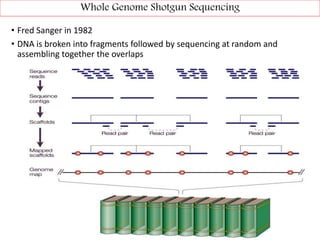
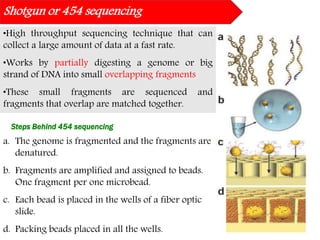
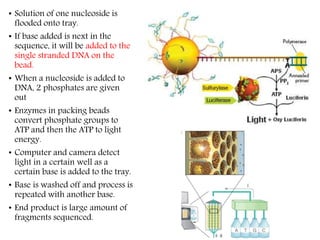
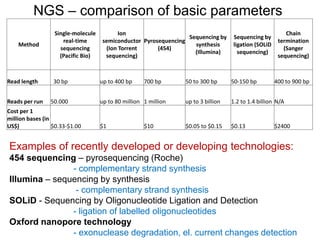
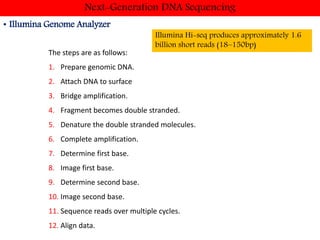
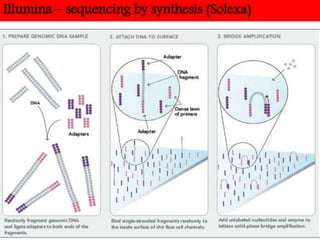
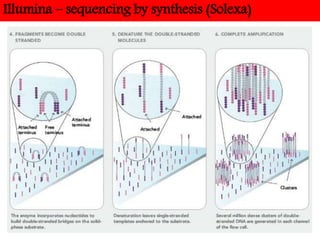
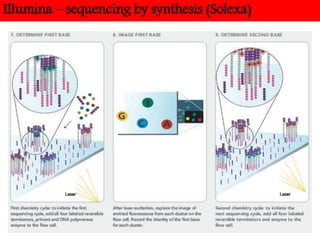
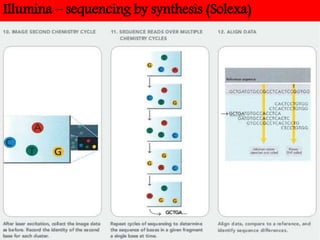
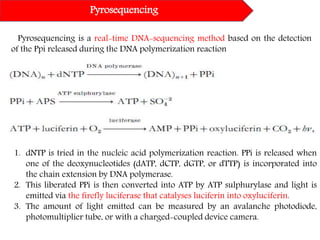
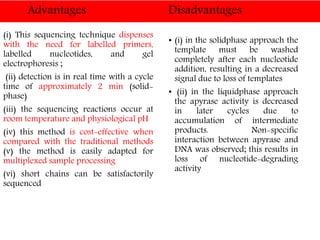
DNA sequencing methods have evolved significantly over time. Early methods like the Maxam-Gilbert chemical method and Sanger chain termination method involved laborious gel electrophoresis. Later developments led to automated Sanger sequencing using fluorescence detection. Next-generation sequencing methods like Illumina sequencing by synthesis and 454 pyrosequencing enabled massively parallel sequencing of many DNA fragments simultaneously. These new methods produce vast amounts of sequence data at lower cost and are used widely in research and applications such as agriculture, medicine, forensics, and more.