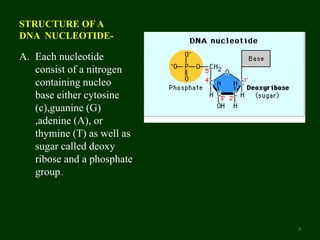
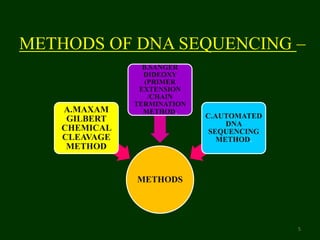
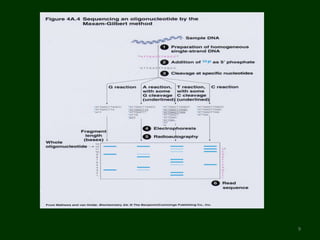
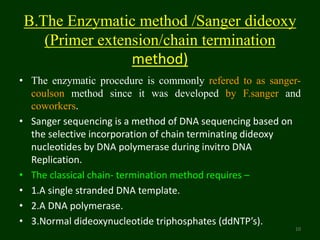
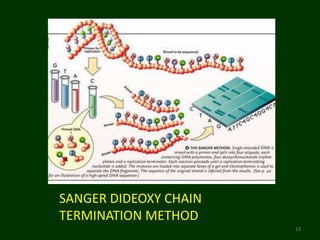
This document provides an overview of DNA sequencing. It begins by defining DNA and its structure. It then explains that DNA sequencing is the process of determining the order of nucleotide bases in a DNA strand. Two common methods of DNA sequencing are described in detail: the Maxam-Gilbert chemical cleavage method and the Sanger dideoxy chain termination method. The document concludes by discussing some applications of DNA sequencing in fields like forensics, medical research, and agriculture.