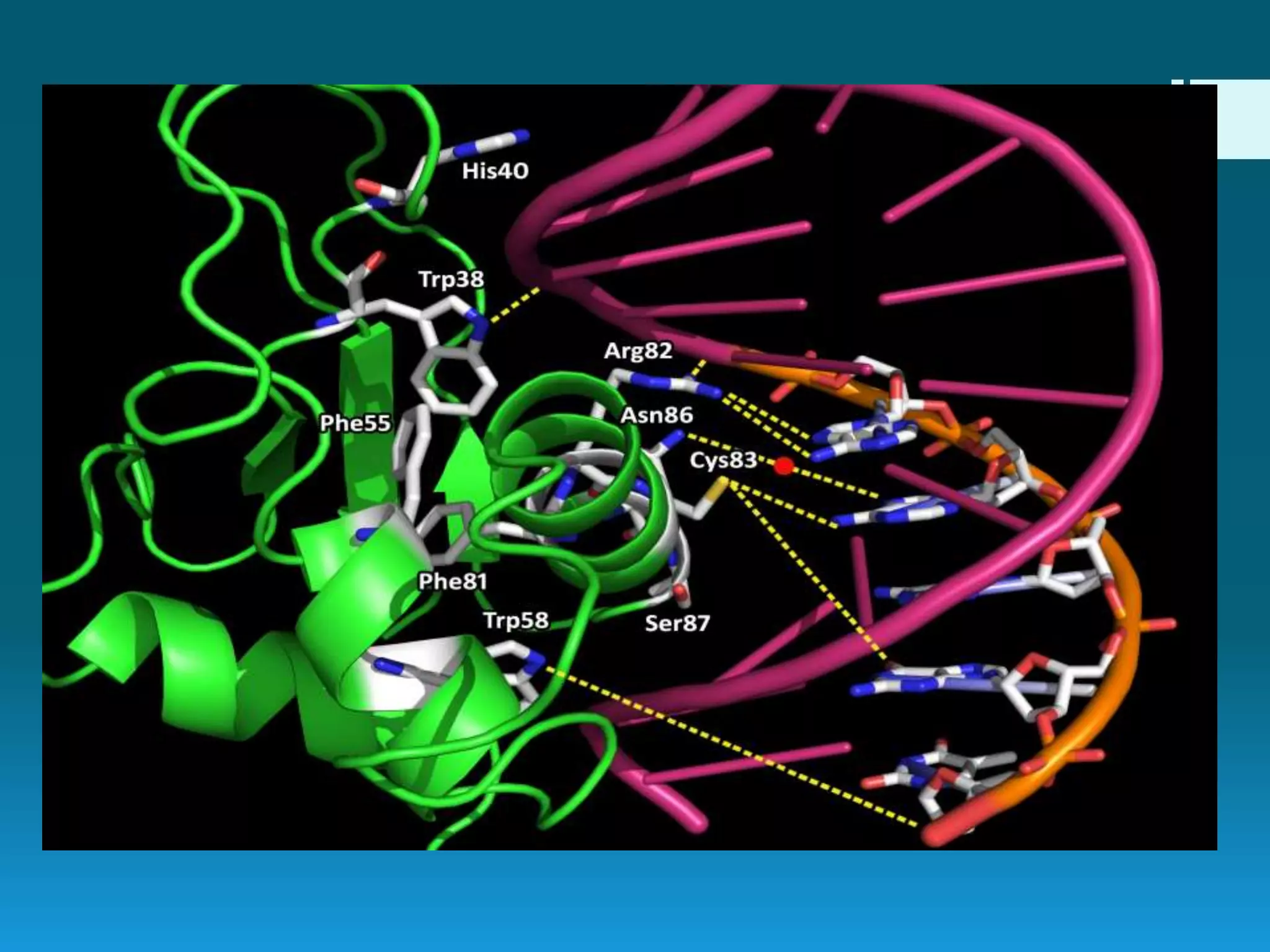
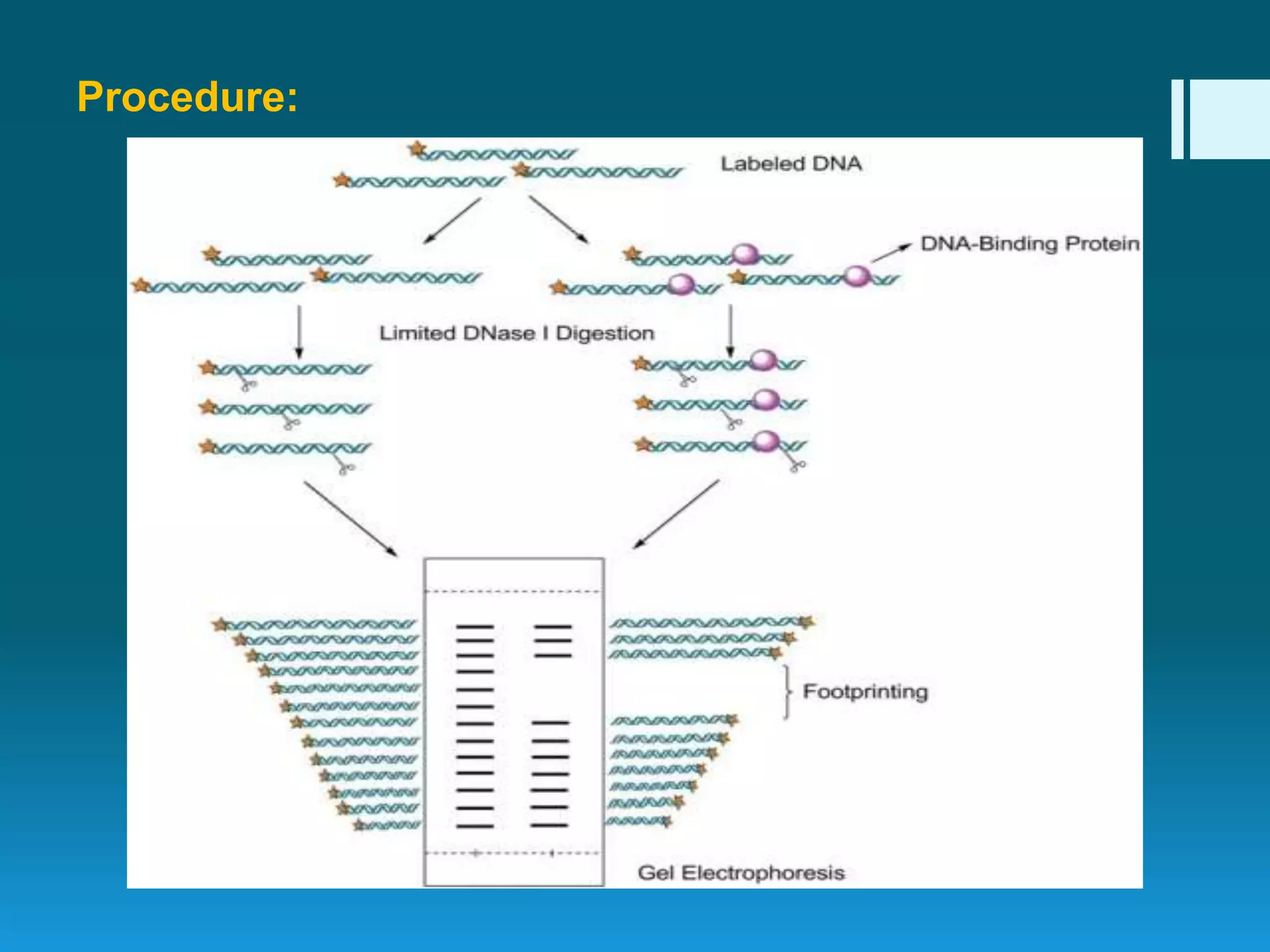
DNA footprinting is a technique used to identify protein binding regions on DNA. It involves treating DNA with nucleases like DNase I, which will degrade the DNA except for regions bound by proteins. These protected regions, called footprints, can identify transcription factor binding sites that regulate gene expression. The technique was originally developed in 1978 to study the binding specificity of the lac repressor protein, and it provides information on DNA-protein interactions and transcriptional regulation.