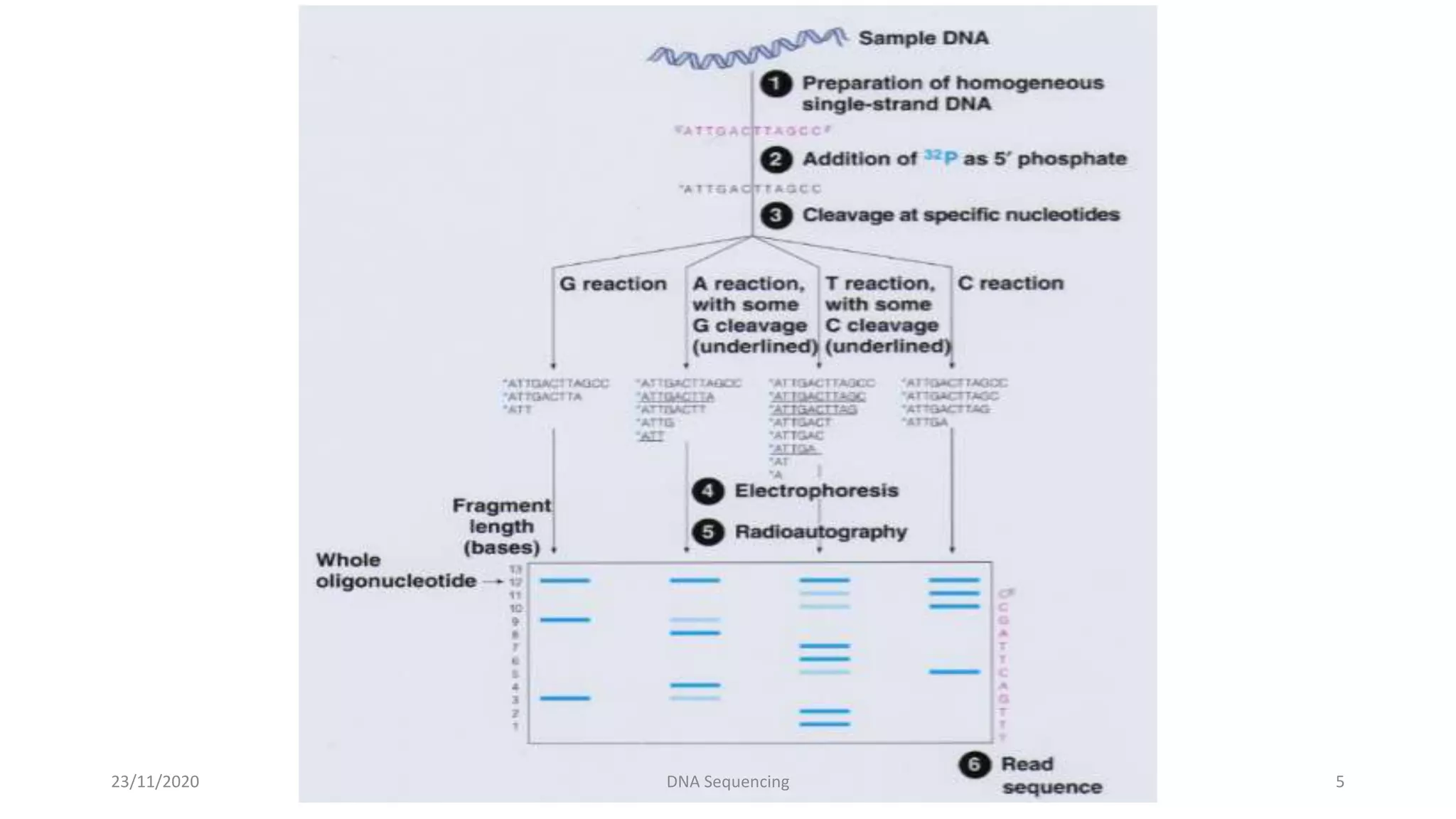
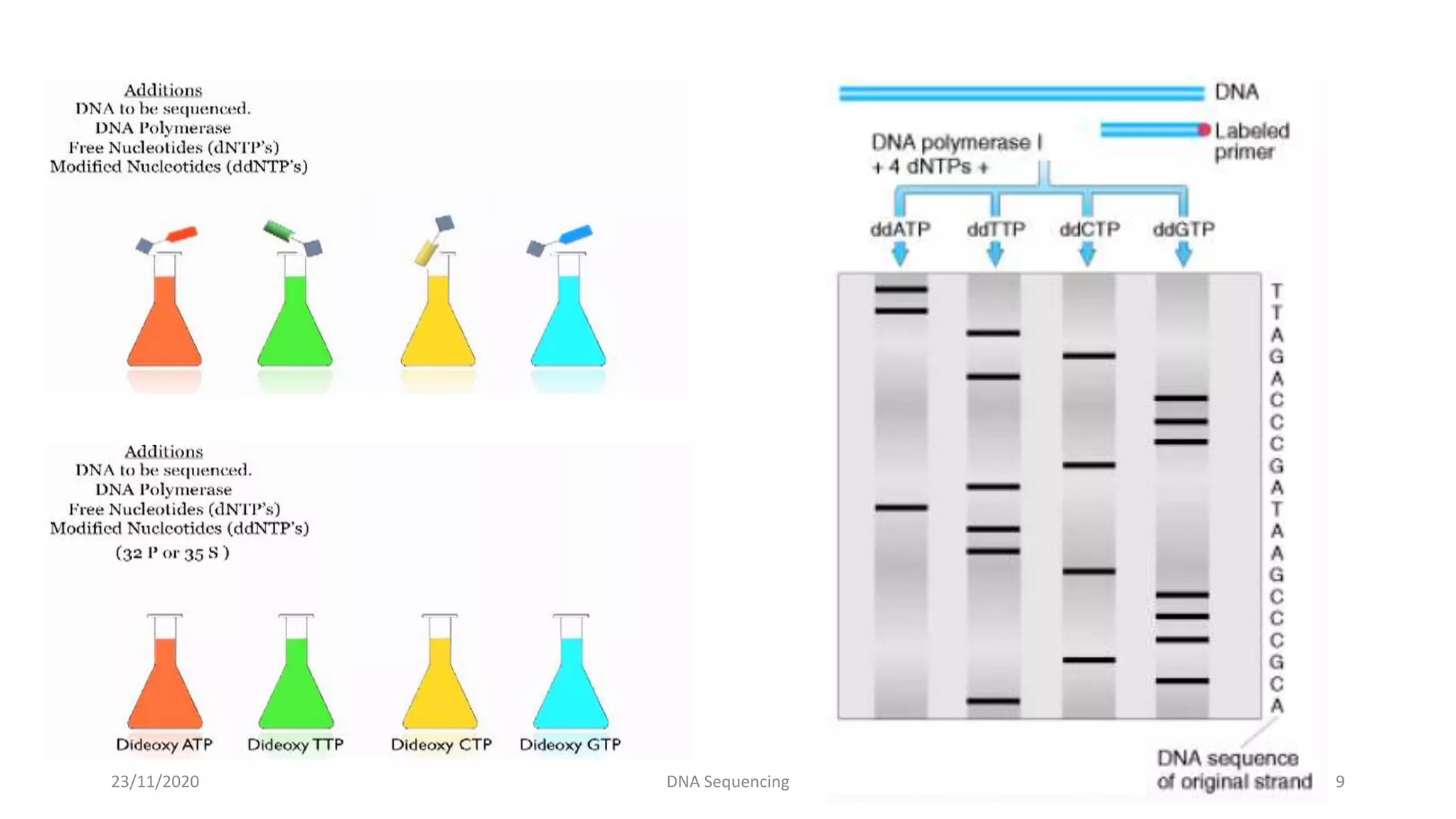
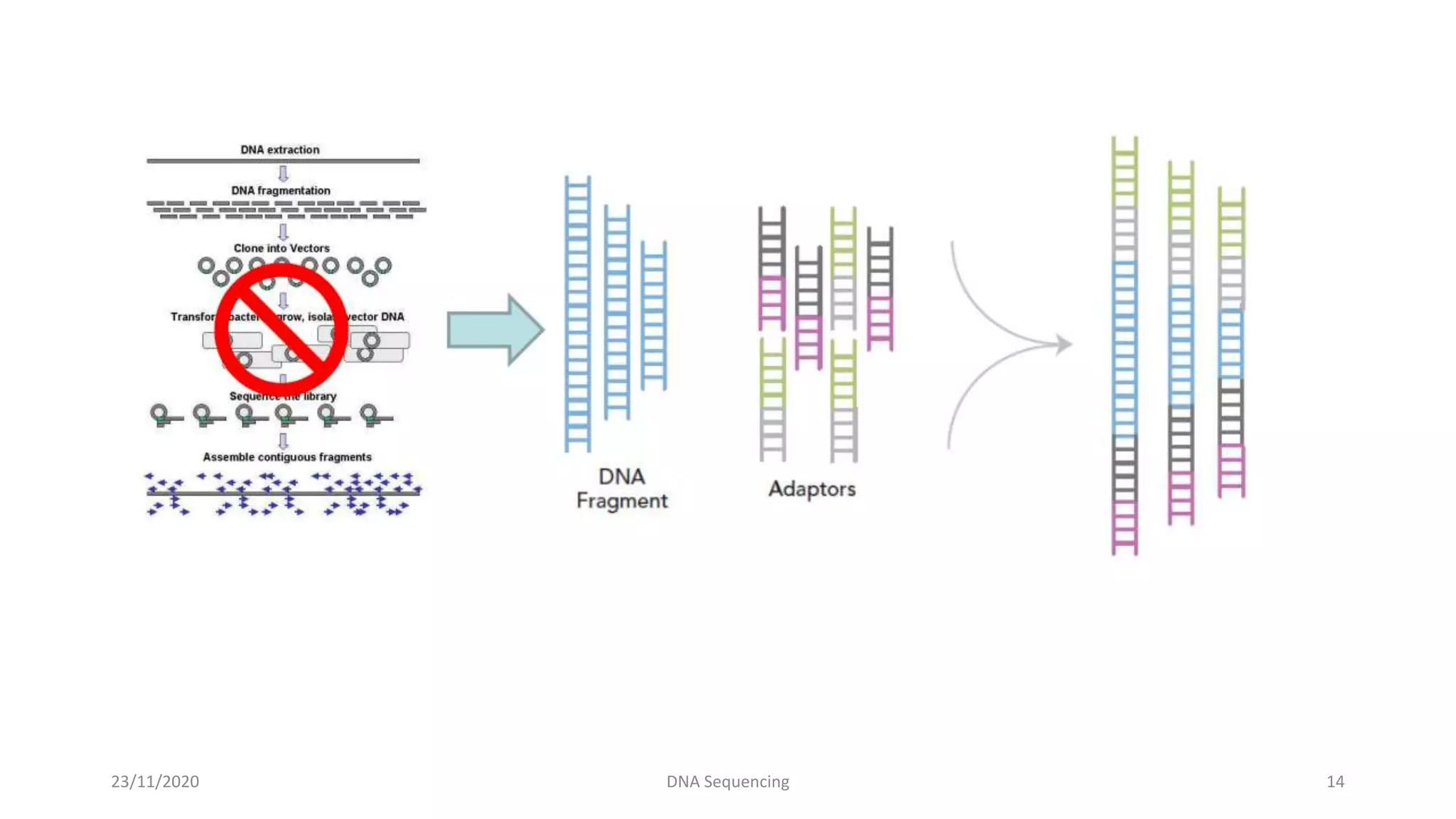
DNA sequencing determines the precise order of nucleotides in a DNA fragment. There are several methods for DNA sequencing, including the chain termination method developed by Sanger, and the Maxam-Gilbert chemical cleavage method. Next generation sequencing is now used, which allows high-throughput sequencing of entire genomes quickly and accurately using automated methods. DNA sequencing has many applications, such as identifying disease-causing genes and mutations.