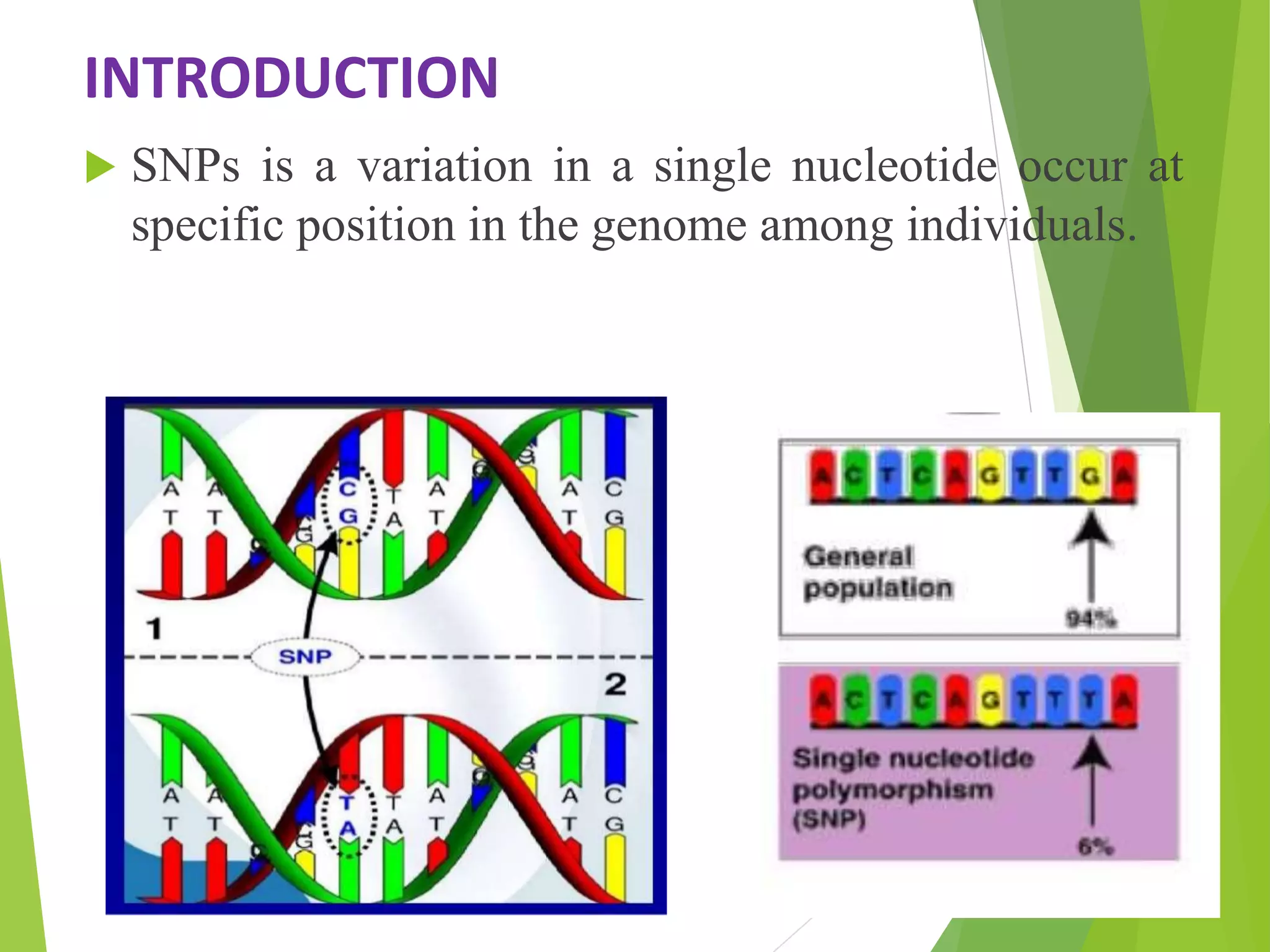
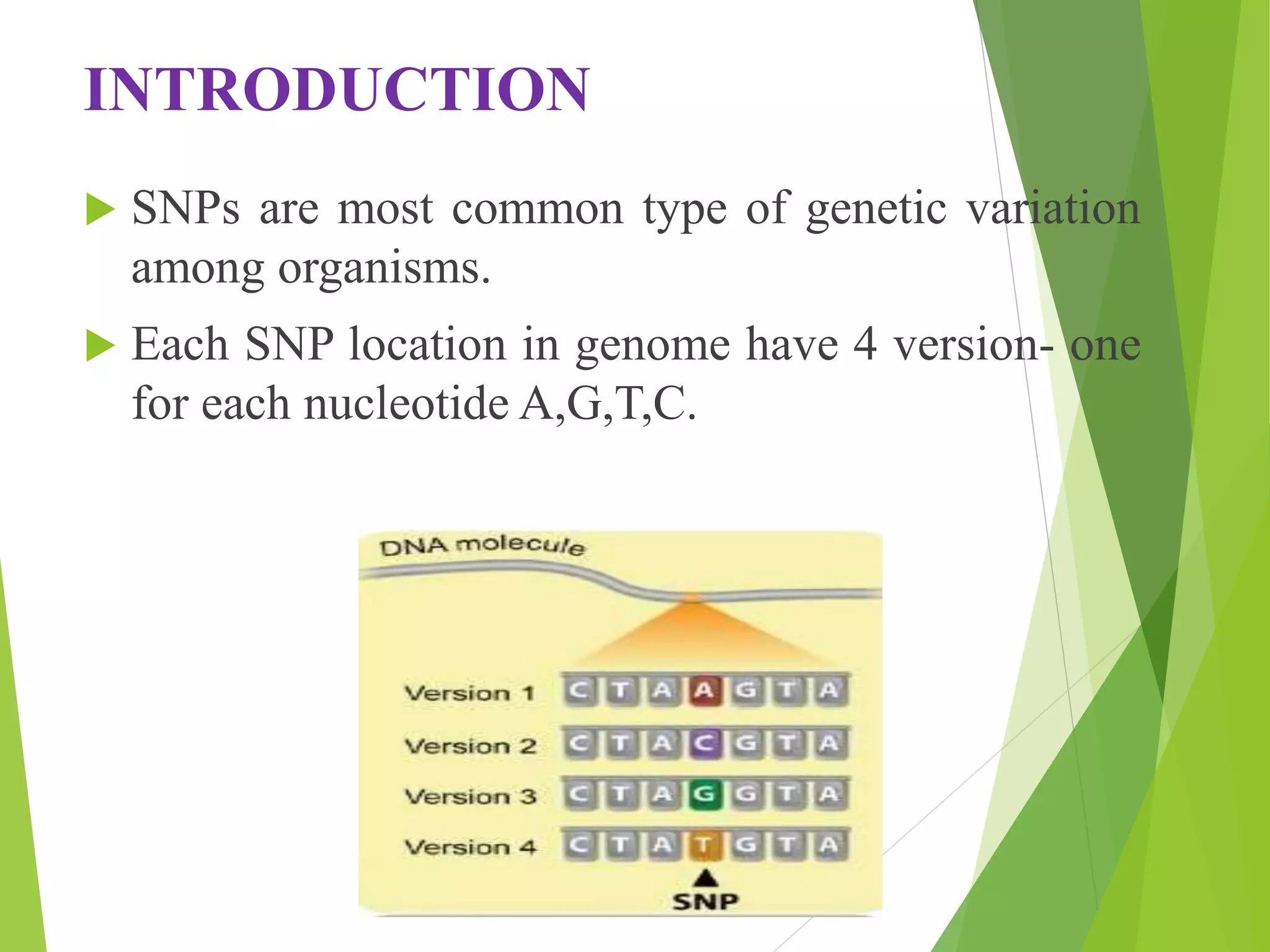
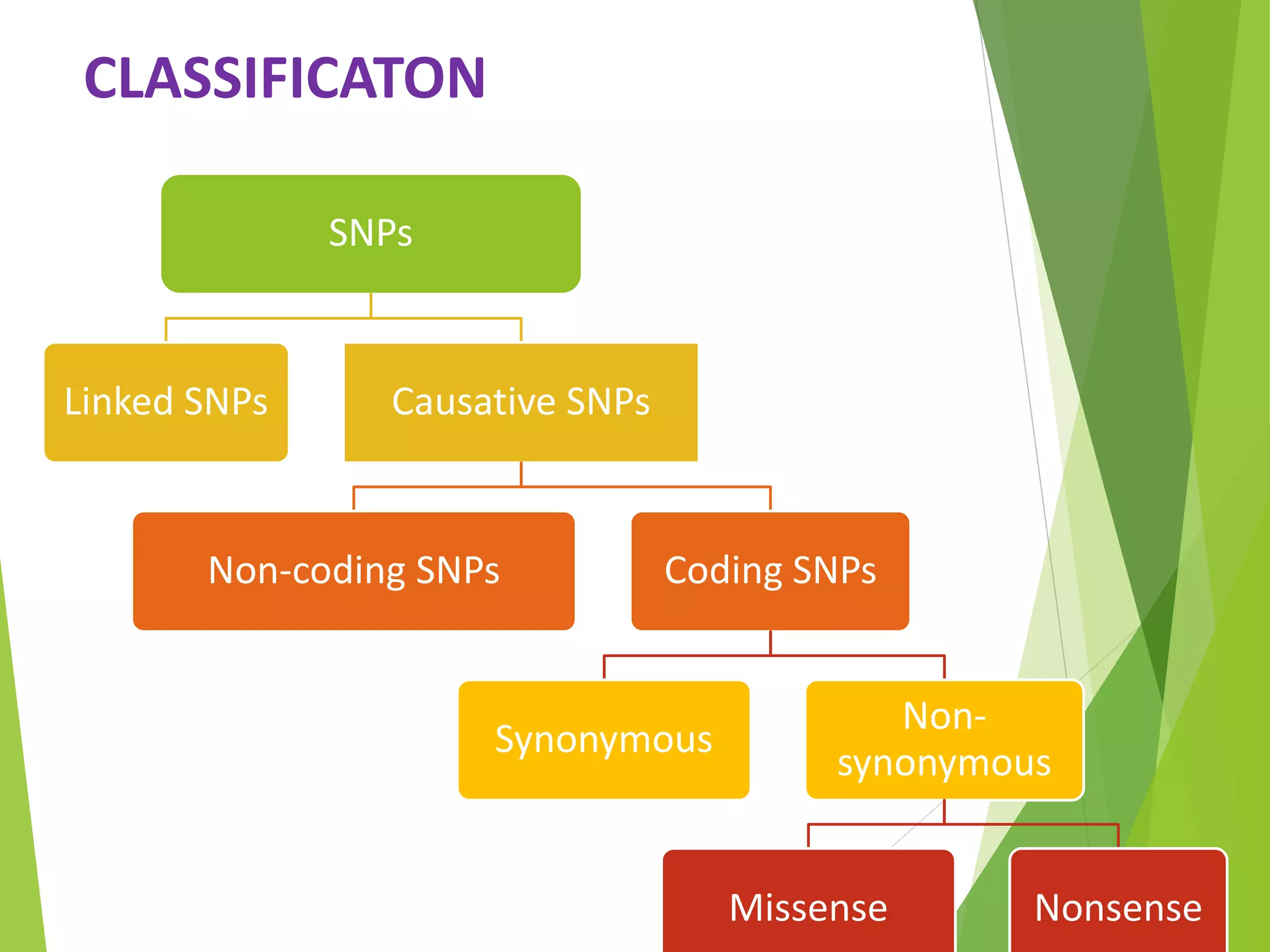
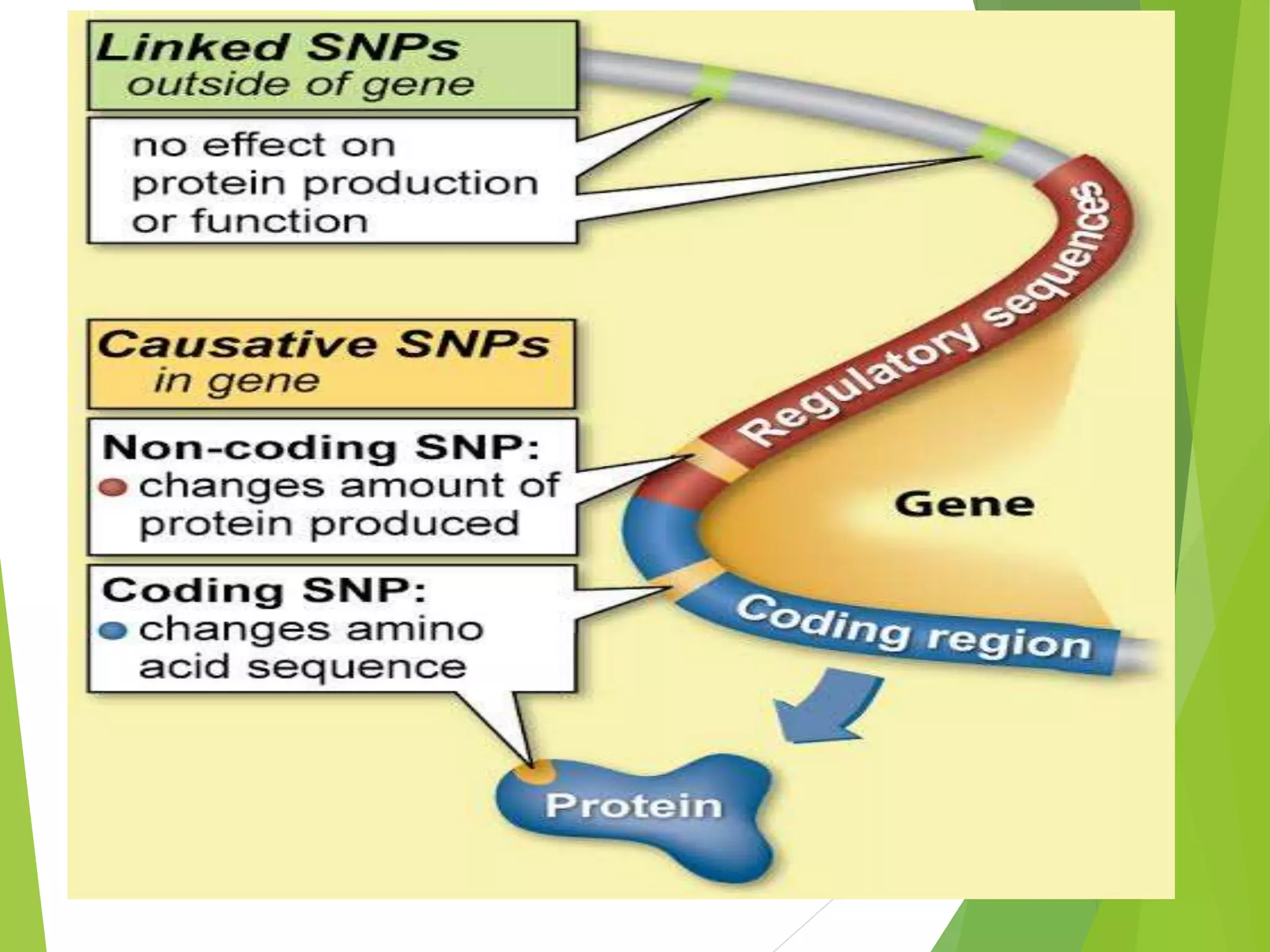
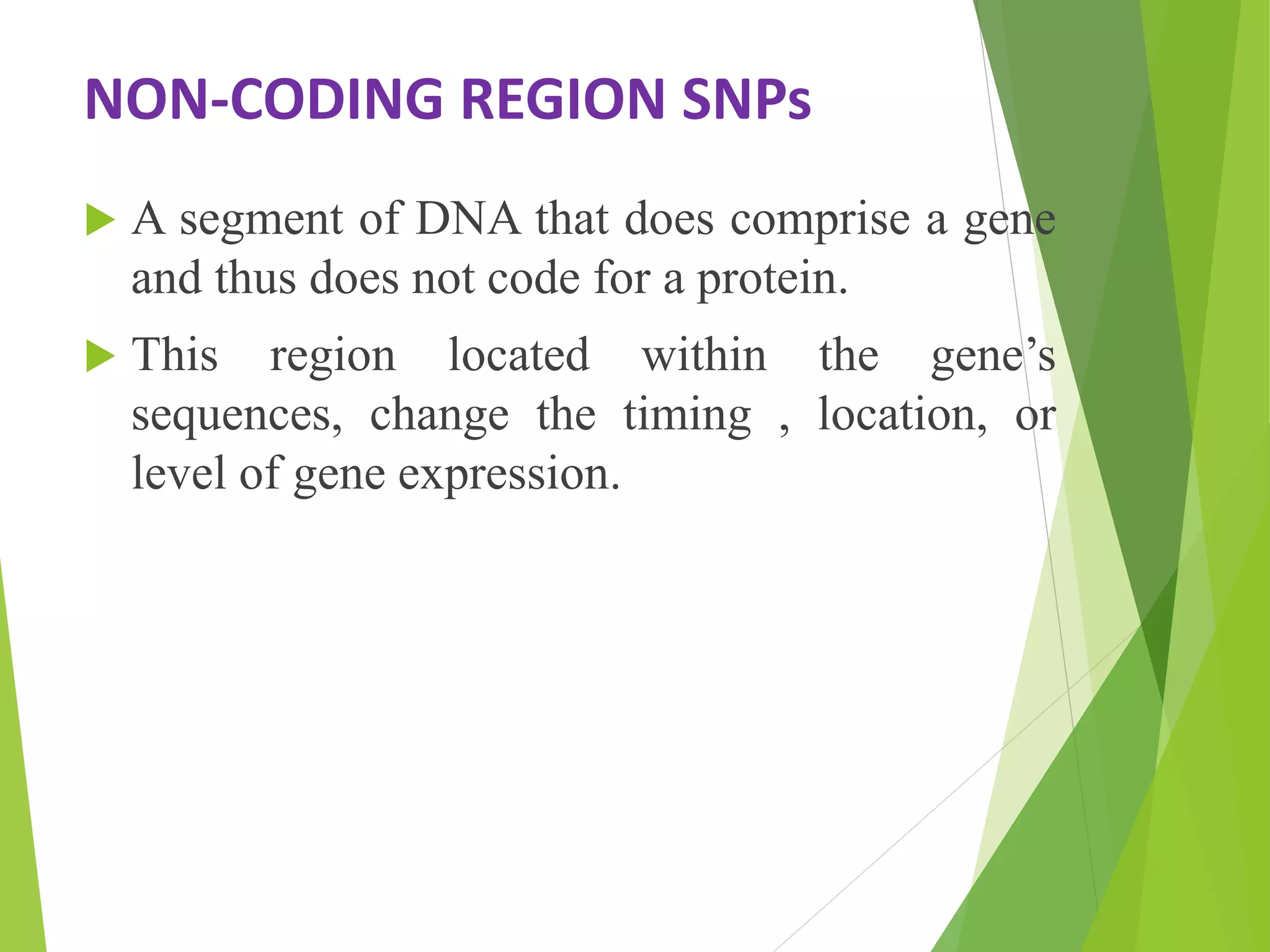
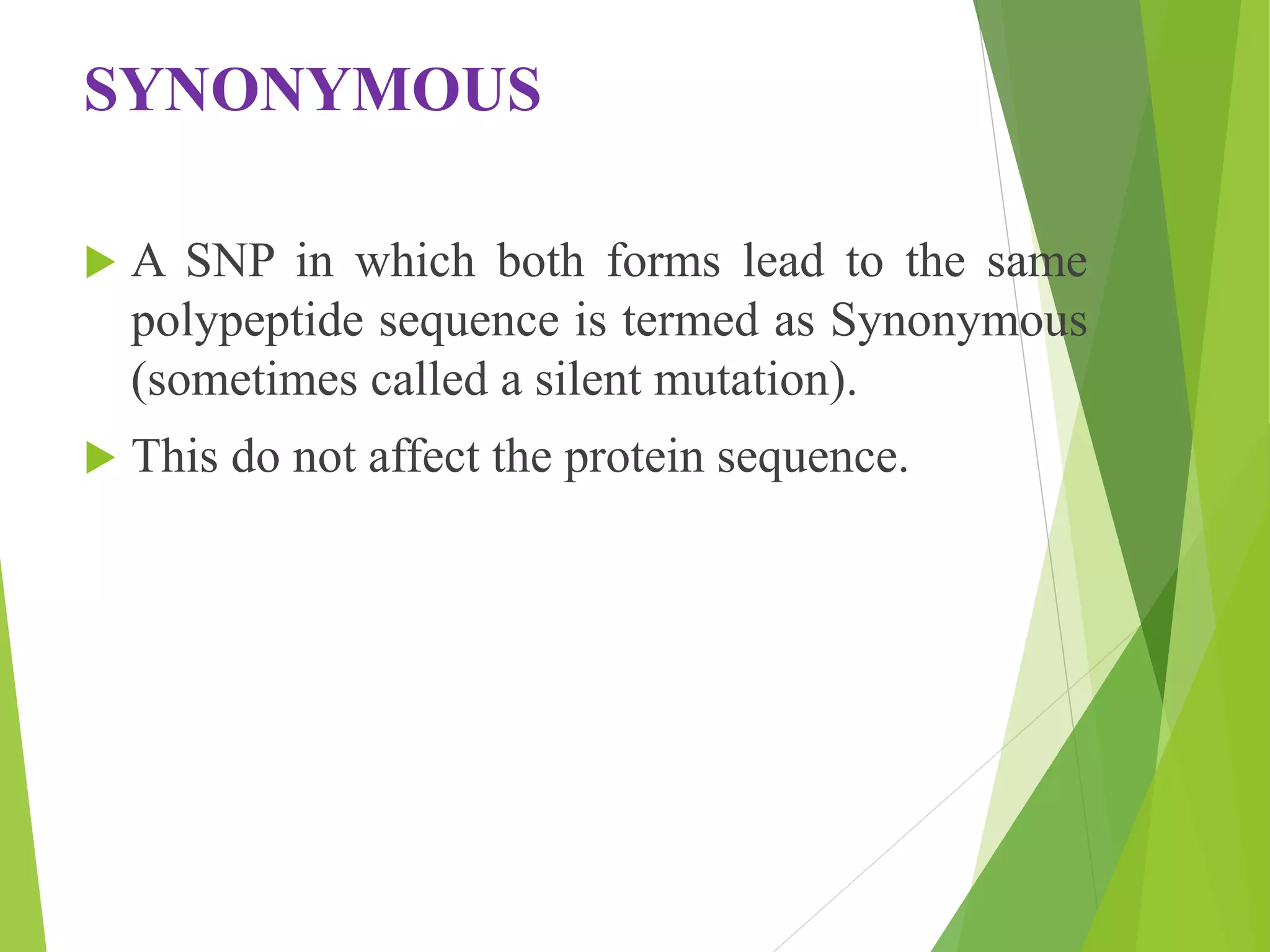
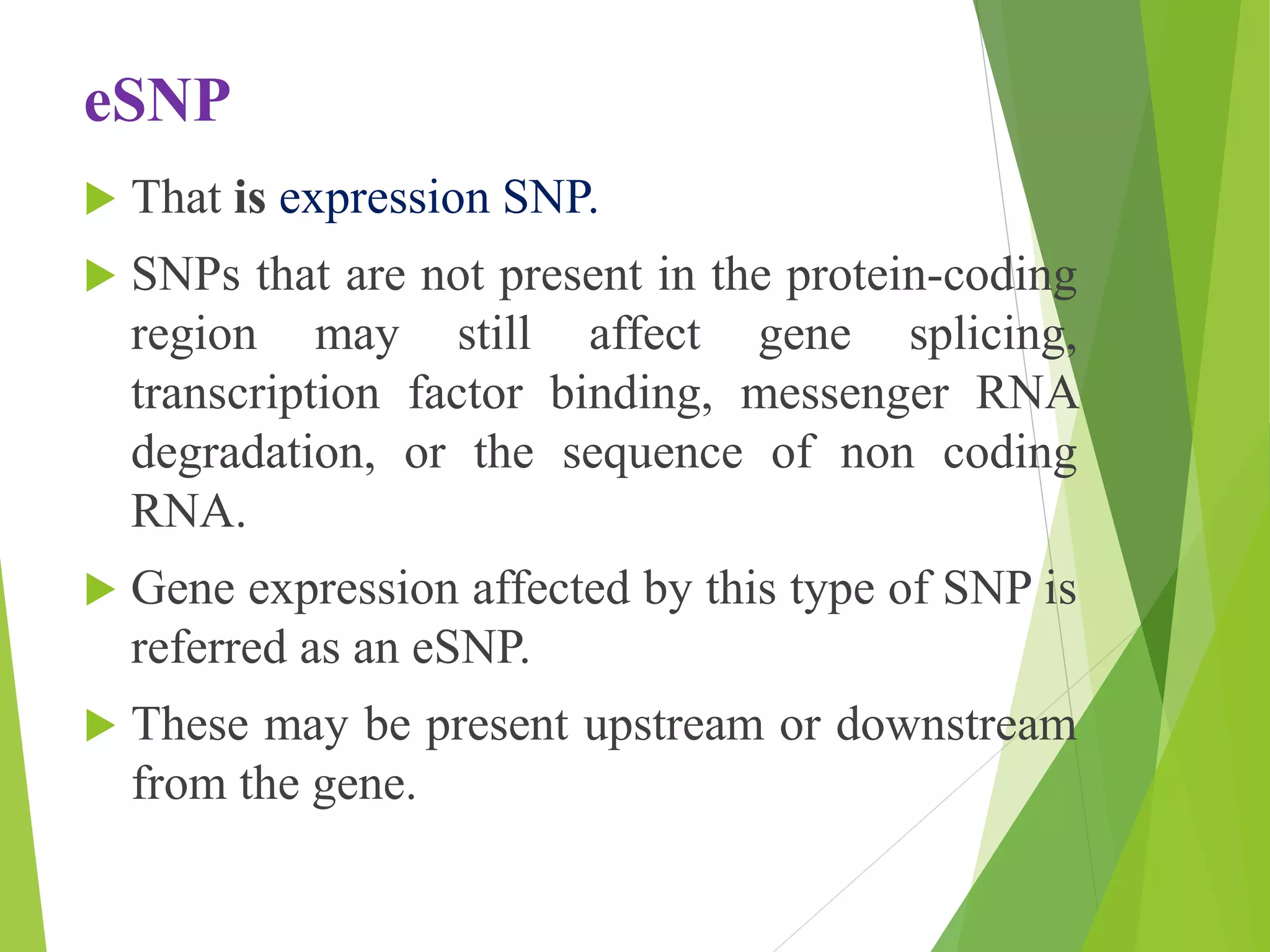
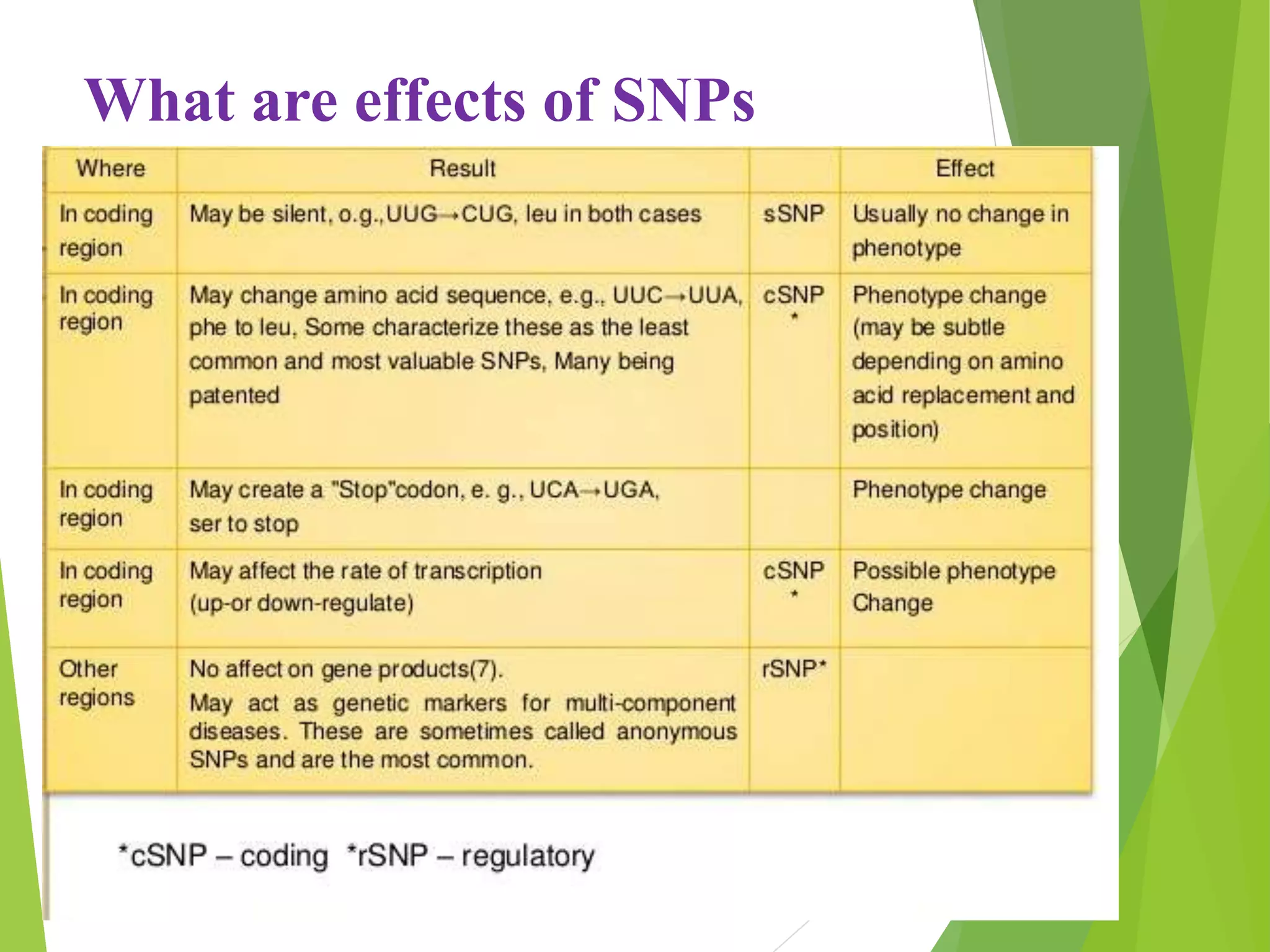
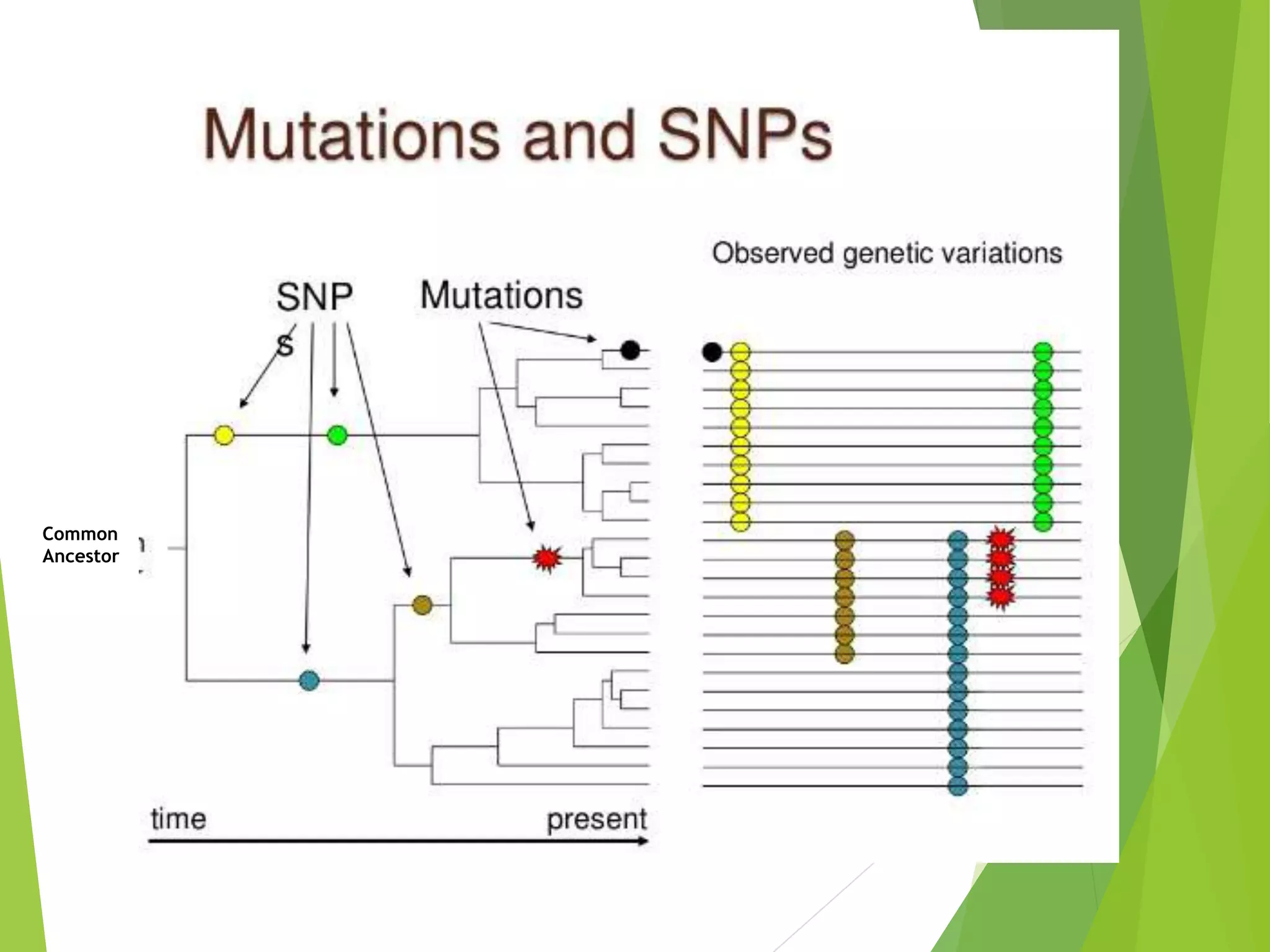
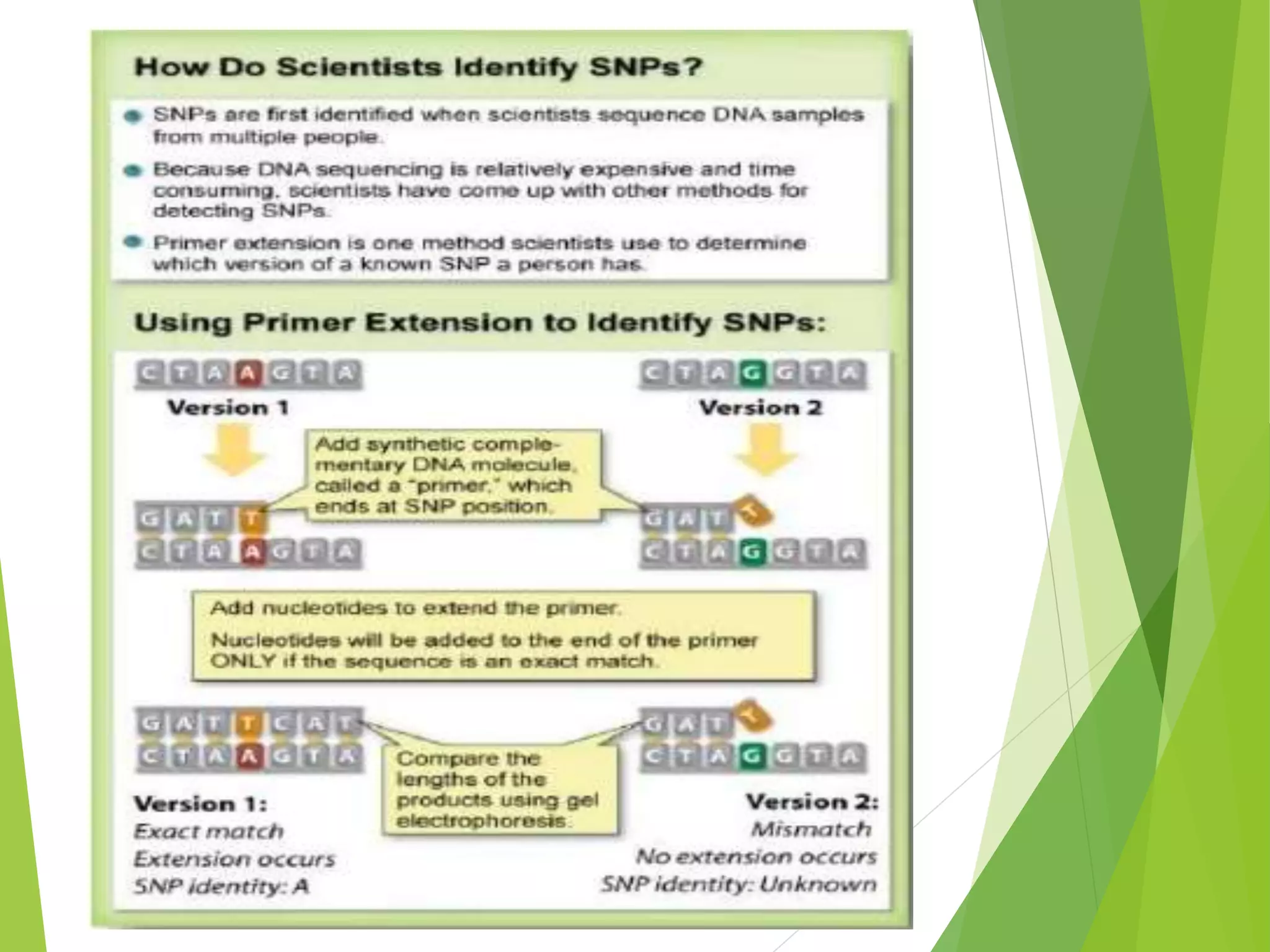
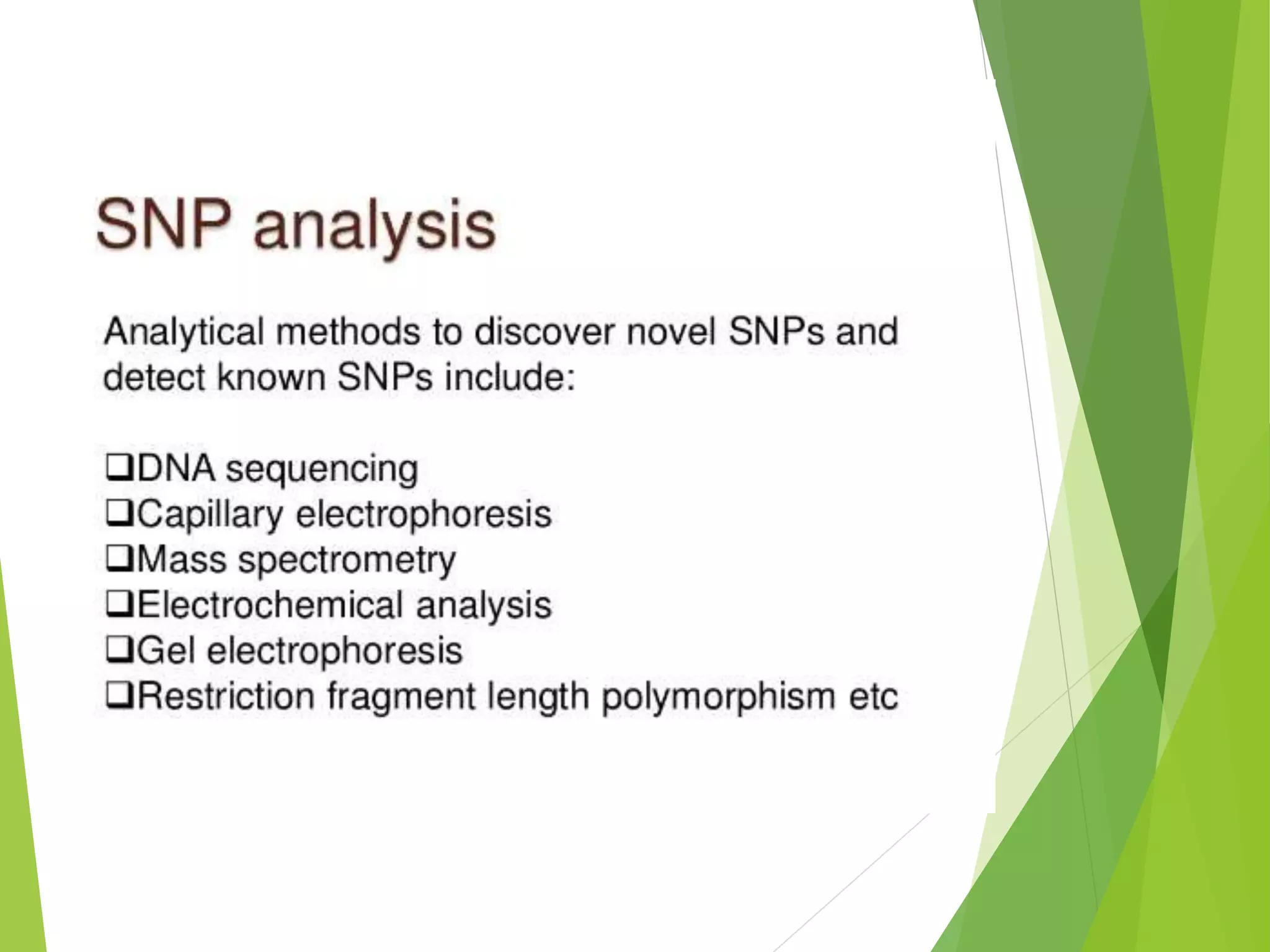
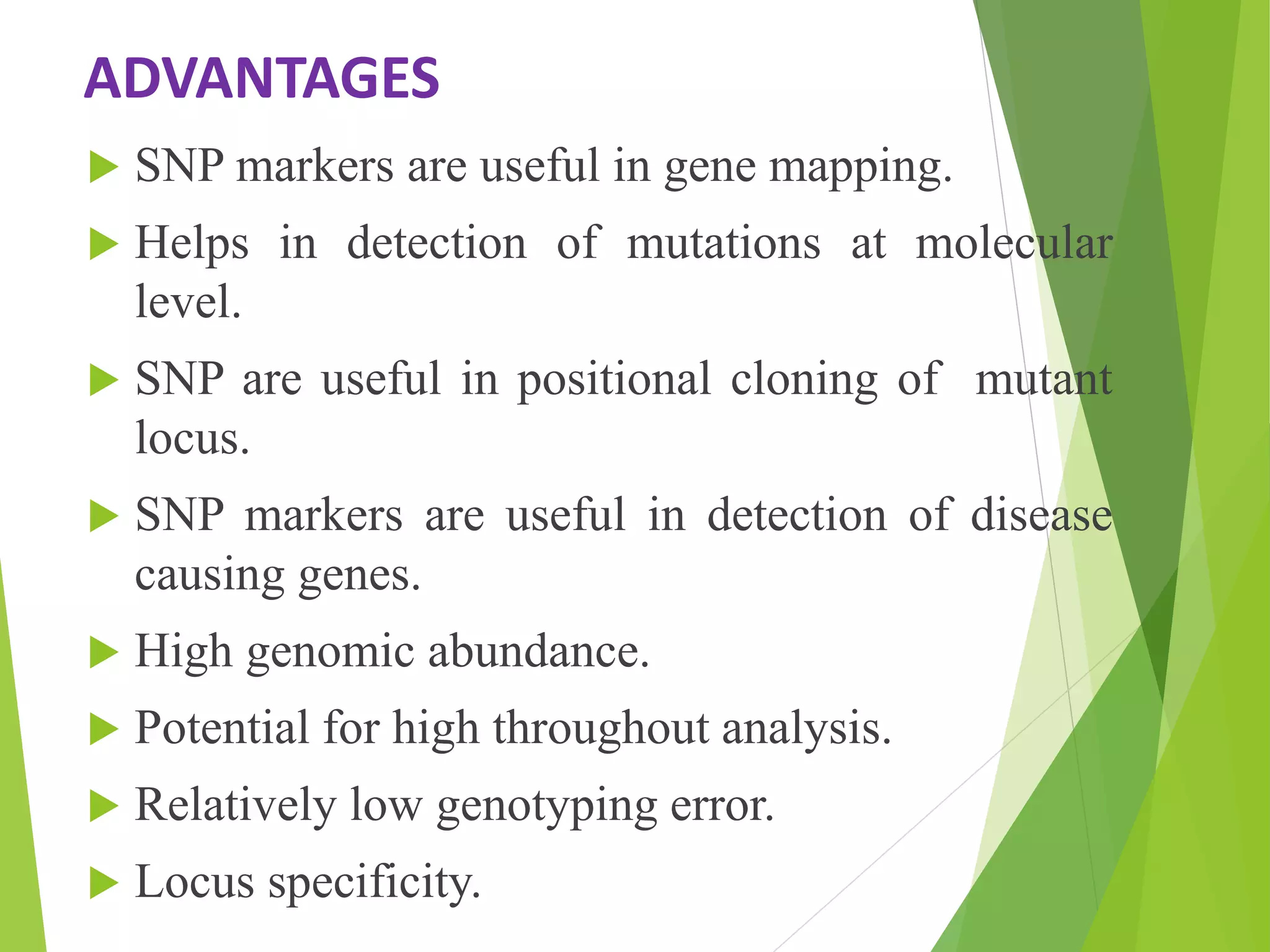
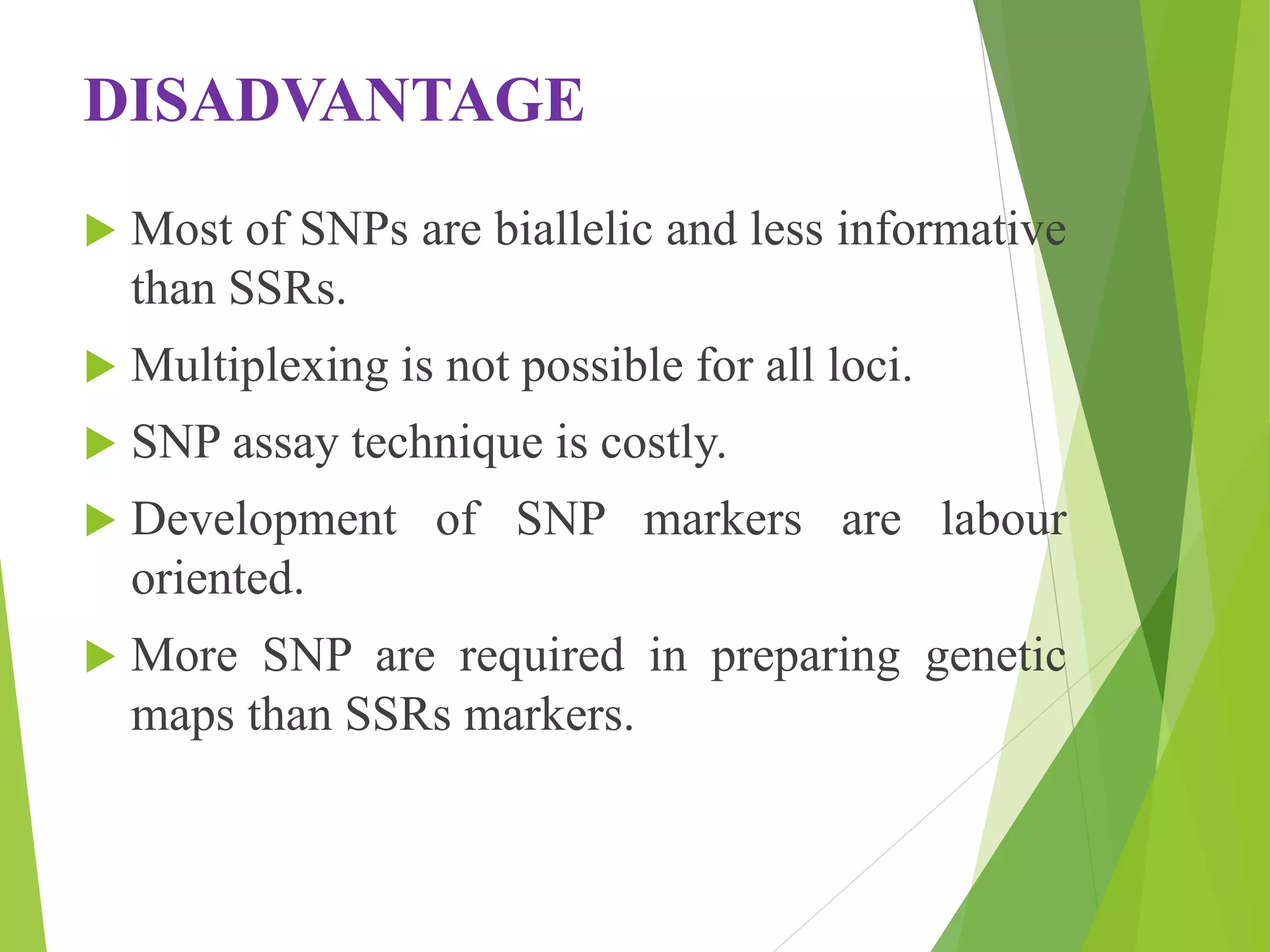
The document discusses single nucleotide polymorphisms (SNPs), highlighting their properties, classification, effects, advantages, and applications in genetics, particularly in livestock. SNPs are the most common genetic variations among organisms and can significantly enhance genetic improvement in cattle and other livestock when used in genetic evaluations and breeding strategies. Additionally, the document covers methods for identifying SNPs and the relevance of SNPs in gene mapping, disease detection, and drug response prediction.