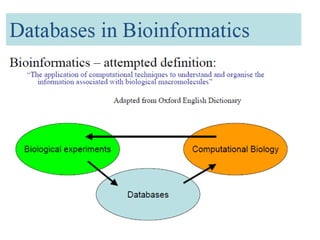
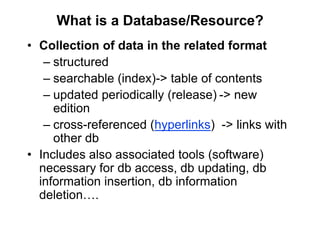
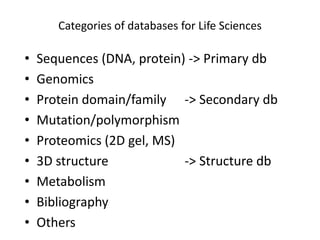
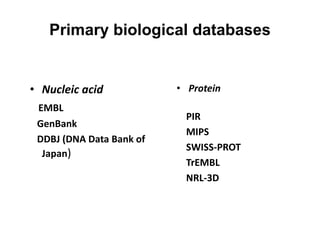
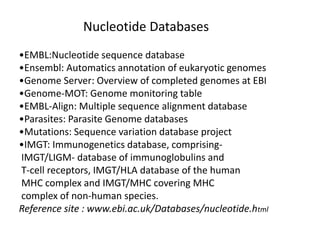
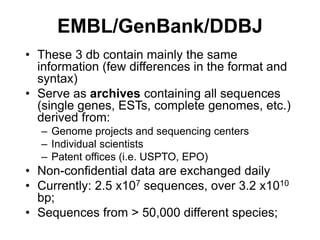
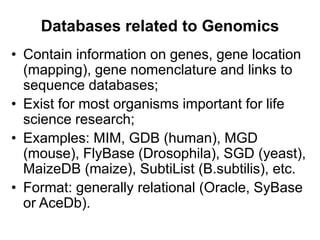
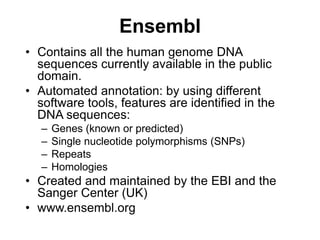
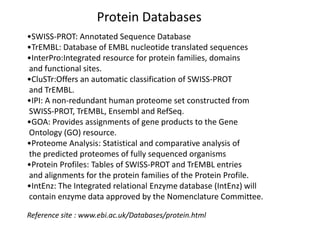
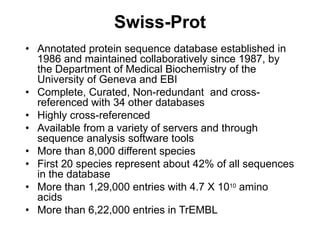
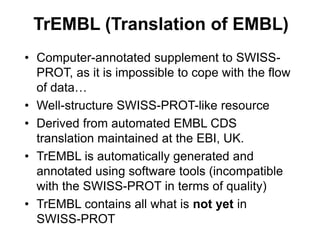
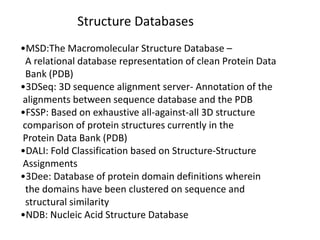
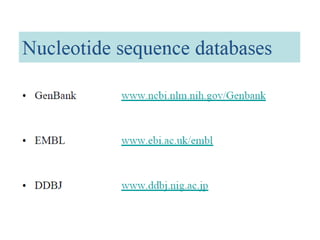
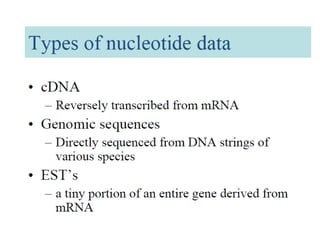
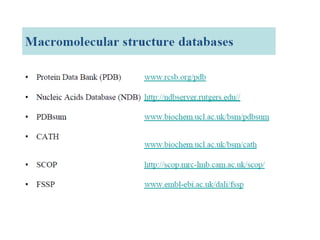
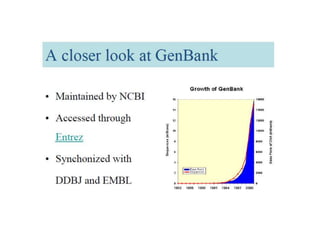
The document provides an overview of various biological databases in bioinformatics, categorizing them into primary, secondary, and integrated databases based on their content and source. It details specific databases for nucleotide, protein sequences, and genomic information, highlighting their functions, examples, and the data they contain. Additionally, it discusses tools for data access and maintenance, including resources like Ensembl, Swiss-Prot, and PubMed.