The document provides an overview of next-generation sequencing technologies, detailing past methods like Sanger and Maxam-Gilbert sequencing, present platforms such as Roche 454 and Illumina, and future developments including Oxford Nanopore sequencers. Each method is described in terms of its operational principles, advantages, disadvantages, and applications. It highlights the evolution of sequencing technologies and their implications for genetic research.

![Sanger Sequencing
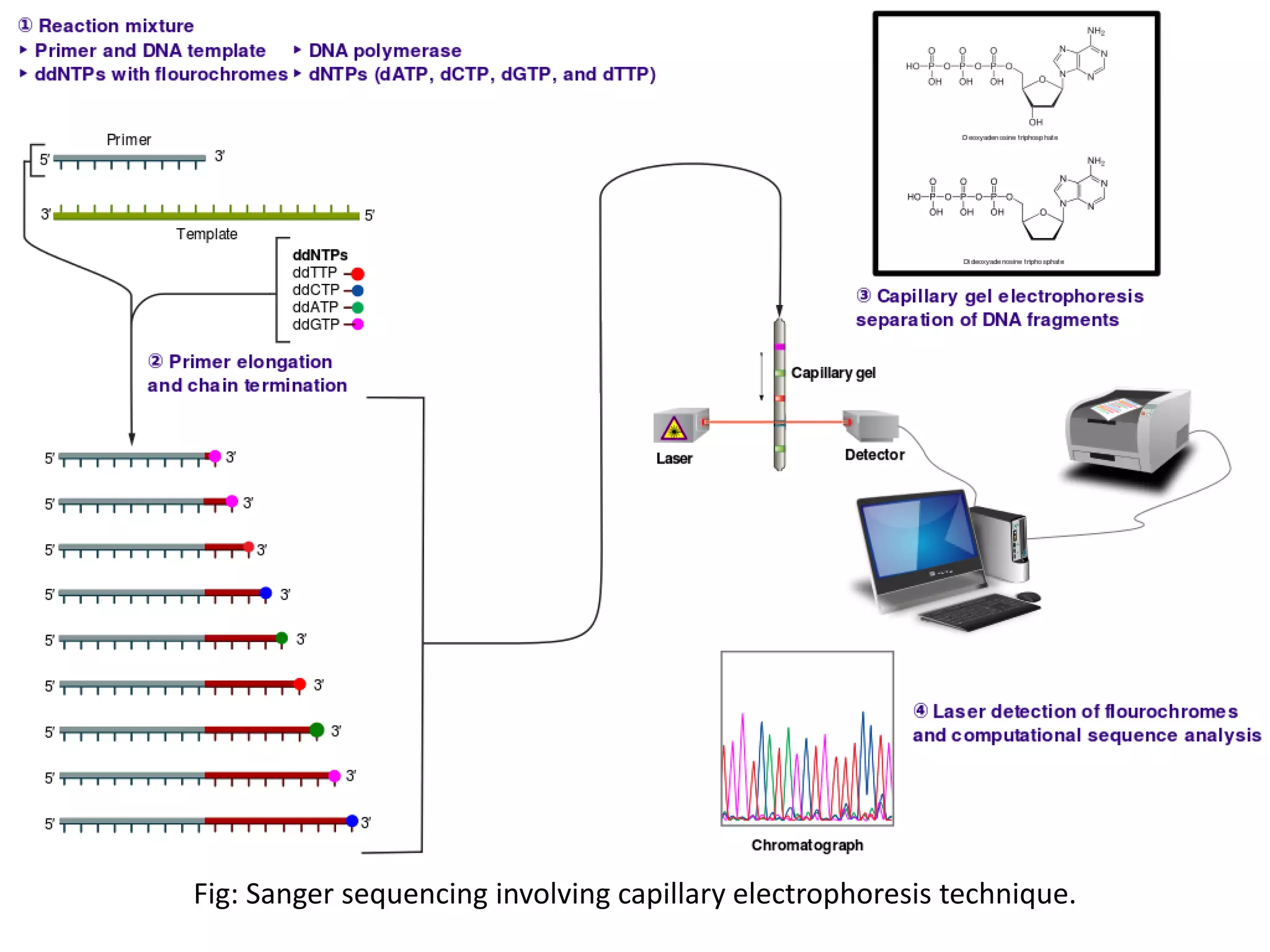
•As we all know, first sequencing technique was developed by Frederick Sanger in 1972. It was
based on chain termination during synthesis[1]. The DNA sequence being synthesized was
terminated using Dideoxynucleotides.
•Sanger sequencing was adopted instead of Maxam-Gilbert Sequencing because of its high
efficiency and low radioactivity as the First generation sequencing.
•AB370: First Automatic Sanger sequencer by Applied Biosystems in 1987 involving Capillary
electrophoresis. Processivity= 96 bases at one time, 500K bases a day with read length up to 600
bases
Deoxynucleotide Dideoxynucleotide](https://image.slidesharecdn.com/nextgenerationsequencing-170219062723/75/Next-generation-sequencing-4-2048.jpg)
![Maxam-Gilbert Sequencing
Another sequencing developed around the same time as that of Sanger was by Sanger’s
colleague Walter Gilbert and Allan Maxam. This sequencing technology was based on chemical
modification of DNA and subsequent cleavage at specific bases[2].
In case of Maxam-Gilbert sequencing[3],
•DNA is cleaved by either Dimethyl sulphate(A/G) or Hydrazine(C/T) in Nucleotide specific
manner and then end labeling is done using 32P Phosphate.
•Partial cleavage at each base produces a nested set of radioactive fragments extending from
the labeled end to each of the position of that base which is then resolved using Polyacrylamide
Gel Electrophoresis.
•Autoradiograph then show band of four different cleavage reaction specific for each base( in a
manner explained by Maxam-Gilbert) and sequence of the DNA fragment can be established by
it.](https://image.slidesharecdn.com/nextgenerationsequencing-170219062723/75/Next-generation-sequencing-6-2048.jpg)
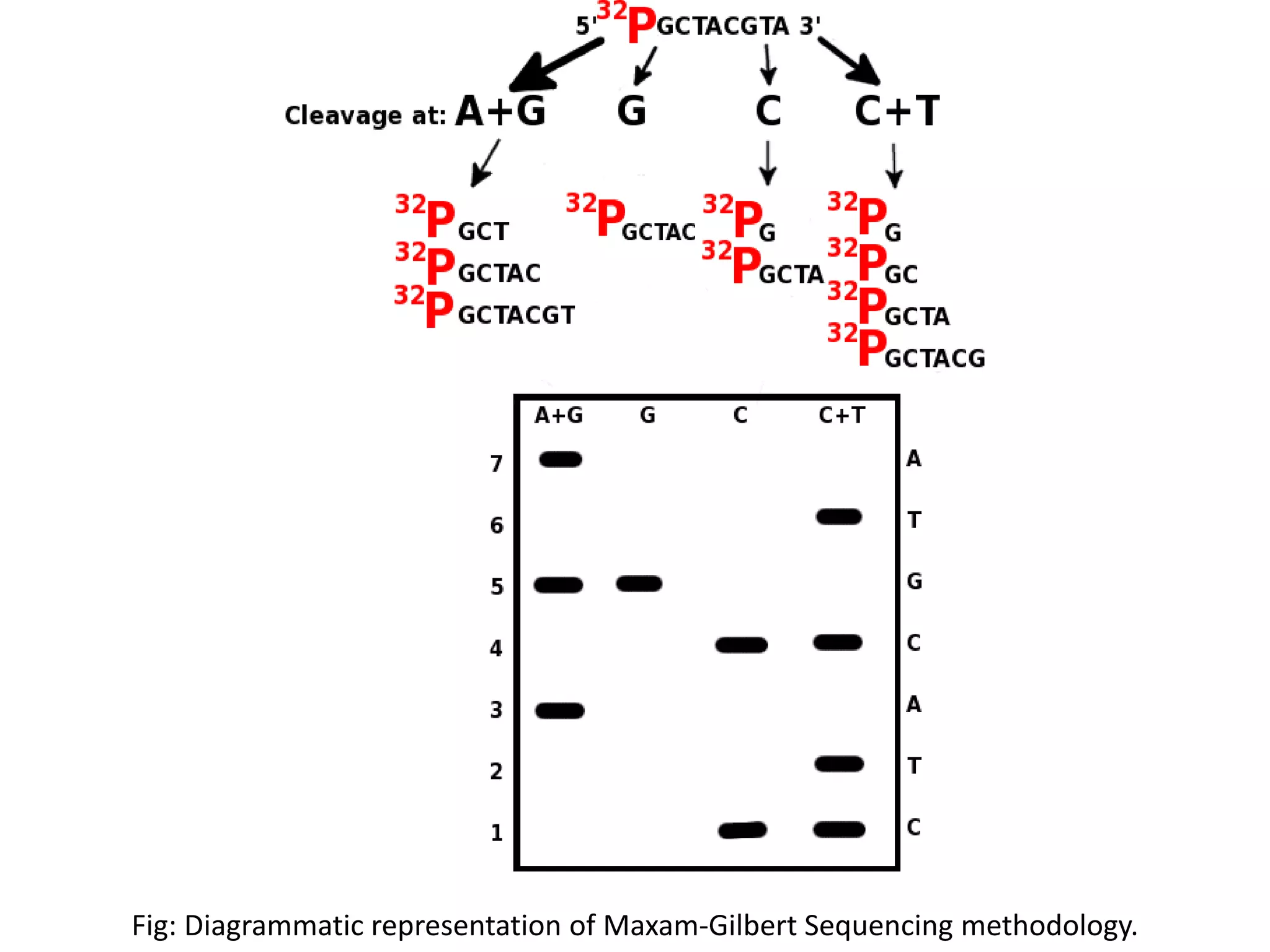
![Fig: Original results of sequencing published by Maxam and Gilbert[3].](https://image.slidesharecdn.com/nextgenerationsequencing-170219062723/75/Next-generation-sequencing-8-2048.jpg)

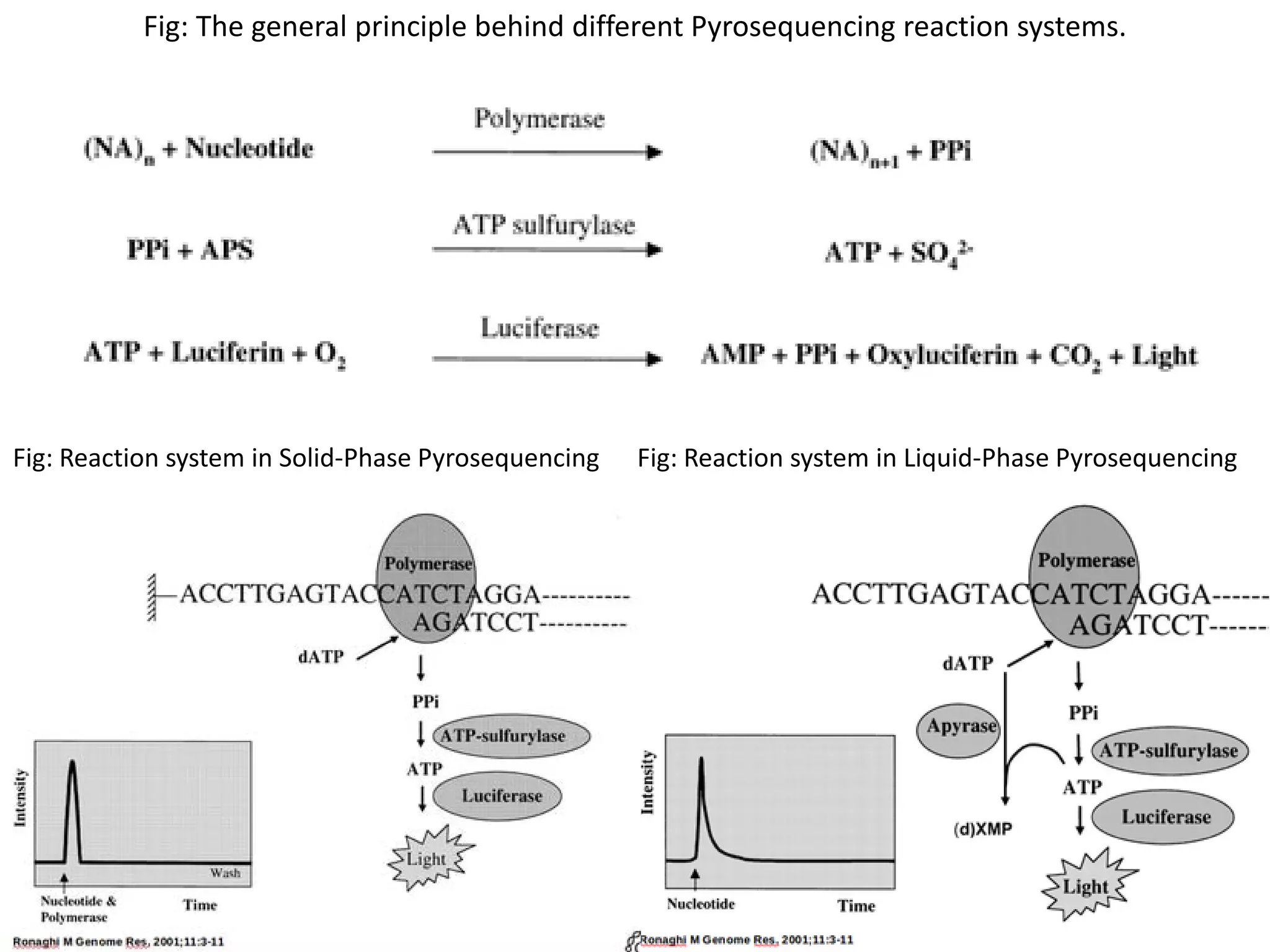
![Roche 454 Sequencer
• Principal: Based on detection of pyrophosphate released during nucleotide incorporation
•In a cascade of enzymatic reactions, visible light is generated that is proportional to the # of
incorporated nucleotides.
•The sequence in which the nucleotides are provided for the reaction in known so the added
nucleotide can be determined by the pyrogram thereby establishing the sequence of the
template DNA sequence.
•Library preparation is majorly same as that of Pyrosequencing.
Enzyme System of 454[4]:
1. DNA Polymerase: Klenow fragment of E.coli DNA Pol1 is used to polymerize daughter strand
from ssDNA template strand.
2. ATP Sulfurylase: ATP sulfurylase used in 454 is a recombinant version from the yeast S.
cerevisiae.
3. Luciferase: Luciferase is from the American firefly Photinus pyralis.
4. Apyrase: It is a nucleotide degrading enzyme from the potato, which is introduced to make a
4 enzyme system. It is used to degrade remaining ATP and dNTPs after every reaction to
prevent non specific or false signals.](https://image.slidesharecdn.com/nextgenerationsequencing-170219062723/75/Next-generation-sequencing-10-2048.jpg)

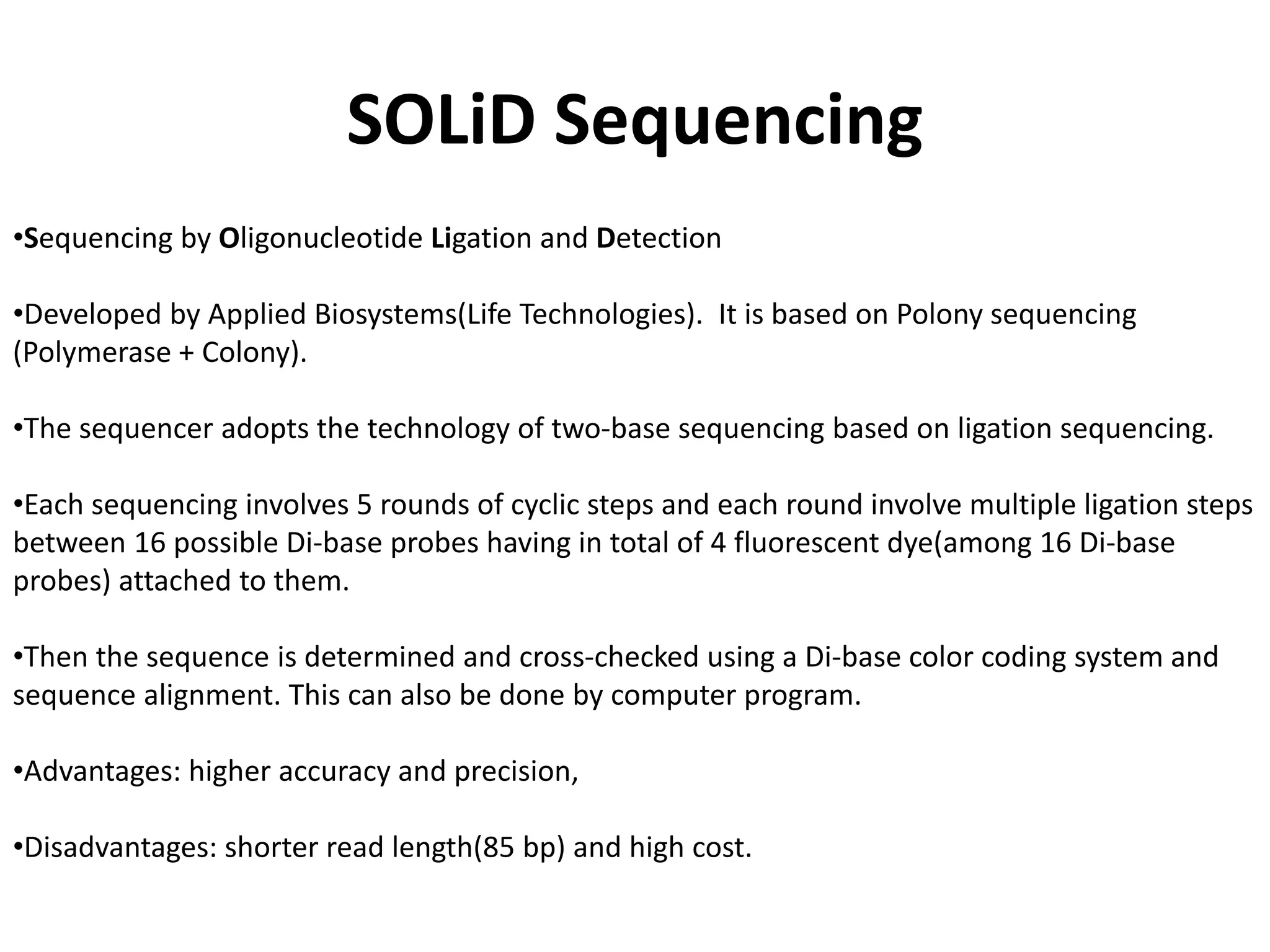
![Appication of SOLiD Sequencing
Application of SOLiD includes[1]:
1. Whole genome sequencing
2. Targeted sequencing
3. Transcriptome research (including gene expression profiling, small RNA analysis, and whole
transcriptome analysis)
4. Epigenome (like ChIPSeq and methylation) analysis.](https://image.slidesharecdn.com/nextgenerationsequencing-170219062723/75/Next-generation-sequencing-14-2048.jpg)
![Probe Anatomy
Fig: Structure of Probe[5]](https://image.slidesharecdn.com/nextgenerationsequencing-170219062723/75/Next-generation-sequencing-15-2048.jpg)
![Fig: Site of cleavage of the probe upon ligation. 5 nucleotides remain attached to the
template while the last 3 along with the fluorescent tag are cleaved off[5].](https://image.slidesharecdn.com/nextgenerationsequencing-170219062723/75/Next-generation-sequencing-16-2048.jpg)
![Results of 1st Round of Sequencing
Problem with running a single round of sequencing is that after round one, fluorescent signals
for every fifth base are known and not the rest, so for that, 4 more rounds of sequencing has to
be performed with primer sequence offset by one base[5].](https://image.slidesharecdn.com/nextgenerationsequencing-170219062723/75/Next-generation-sequencing-17-2048.jpg)
![Primer Specification for Every Round
For every subsequent round, the sequence of primer offsets by one base[5].](https://image.slidesharecdn.com/nextgenerationsequencing-170219062723/75/Next-generation-sequencing-18-2048.jpg)
![Di-Base Colour coding system
Fig: Di-base colour coding system developed for SOLiD sequencing only [5].](https://image.slidesharecdn.com/nextgenerationsequencing-170219062723/75/Next-generation-sequencing-19-2048.jpg)
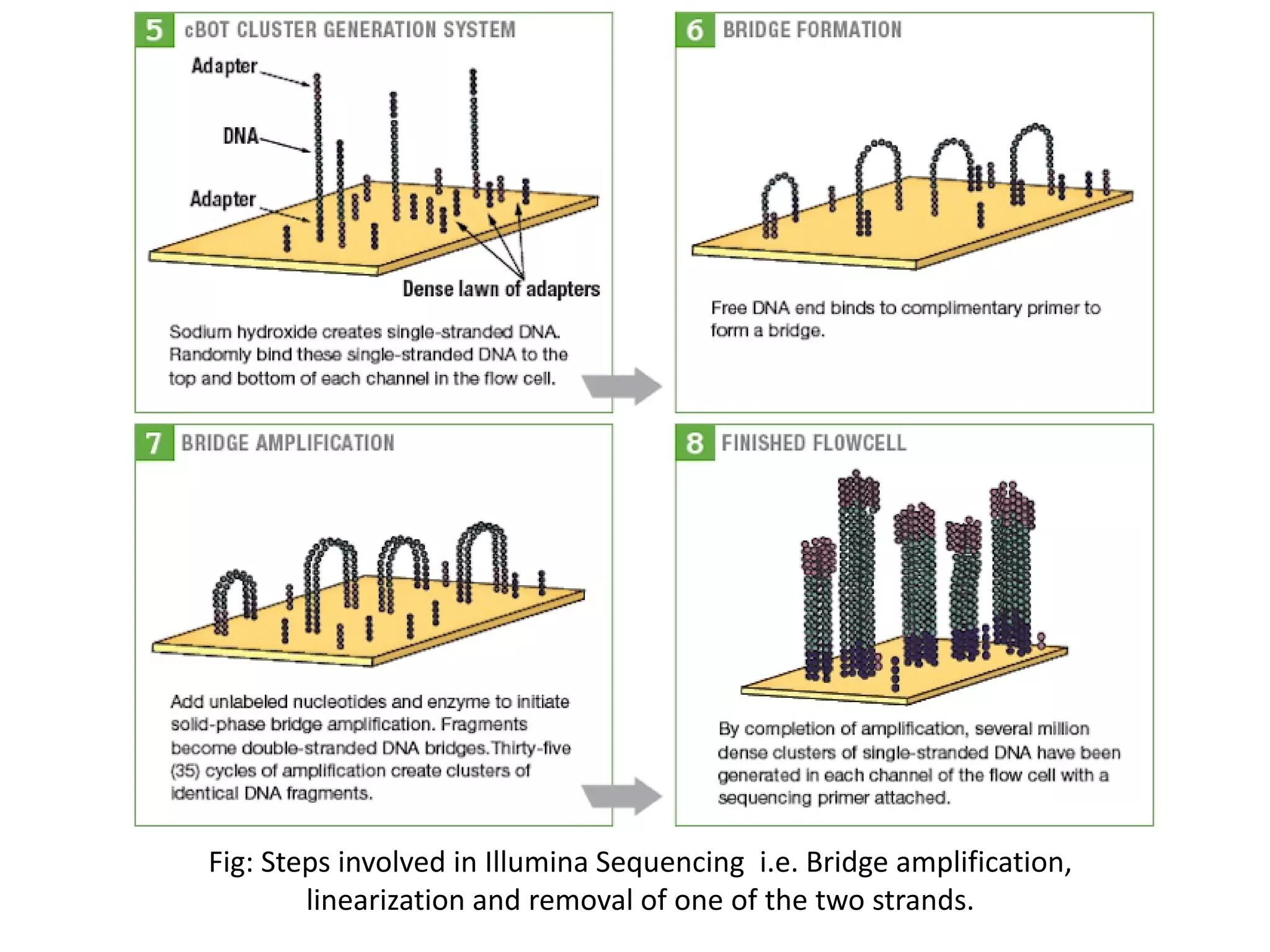
![Illumina- Sequencing by Synthesis
•Sequencing by Synthesis is the proprietary method of Illumina sequencers and involves Bridge
Clonal Amplification of one of the strand after hybridisation to the flow cell surface.
•Hybridisation takes place with the help of one of the adaptors attached to the template.
•After clonal amplification, again polymerase start synthesizing the strand but this time
nucleotides complexed with fluorescent tag are incorporated into the strand and the fluorescent
tag is recognized by the detector. In this way sequence of both forward and reverse strand is
obtained and differentiated by index sequence specific for both forward and reverse strand[6].
Fig: Bridge Amplification
during sequence by
synthesis](https://image.slidesharecdn.com/nextgenerationsequencing-170219062723/75/Next-generation-sequencing-20-2048.jpg)

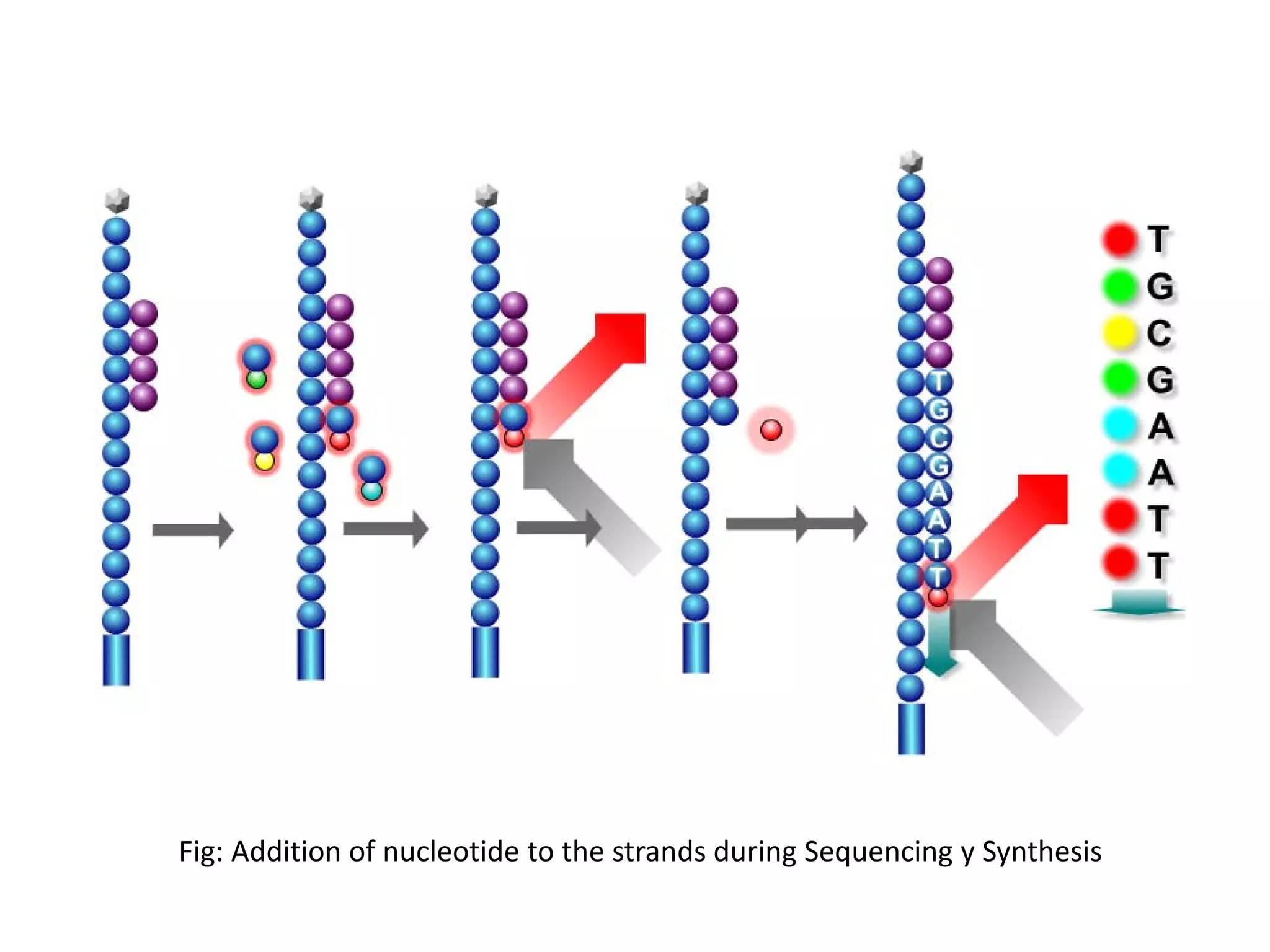

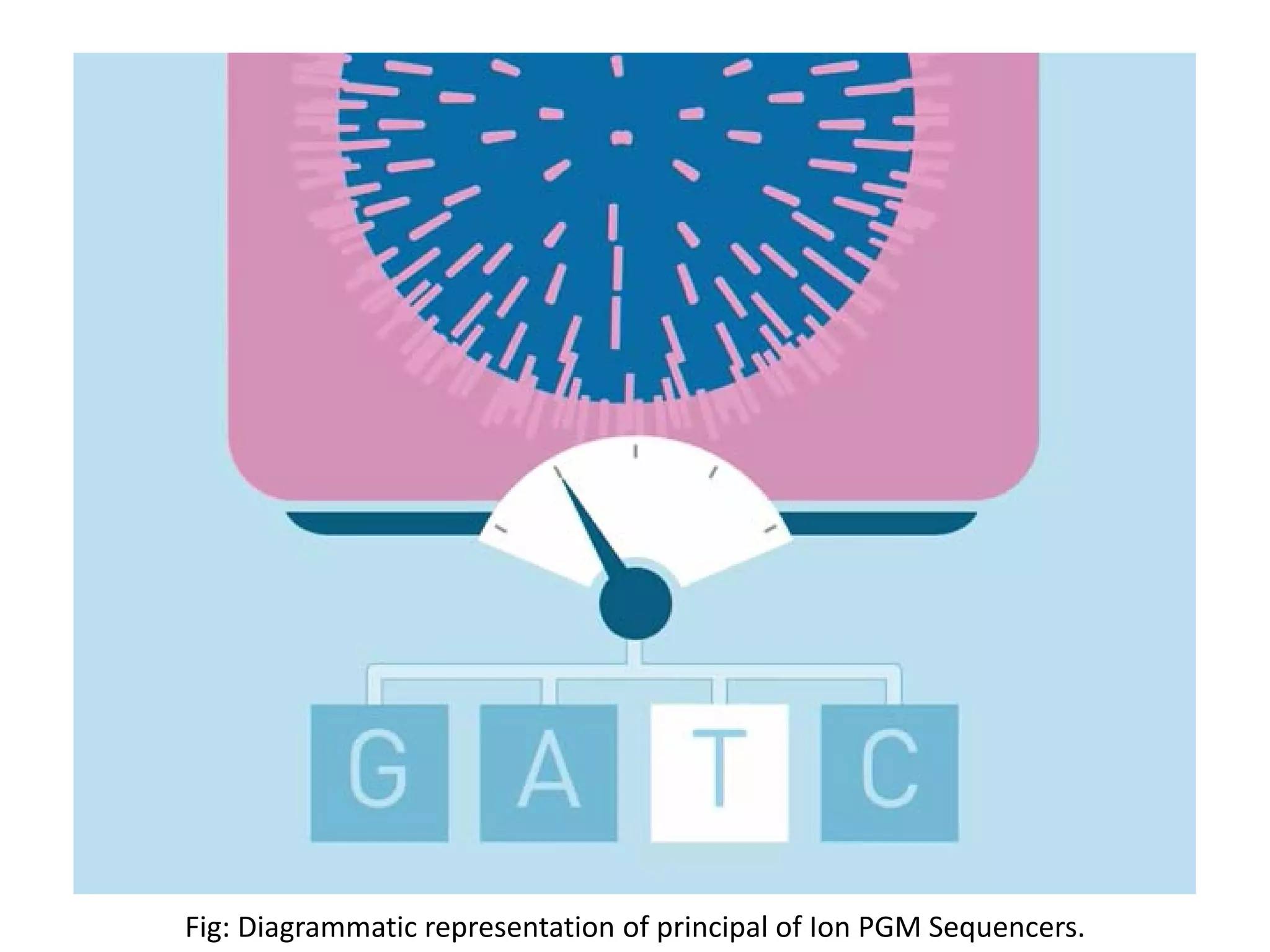
![ION PGM Sequencers
•Ion PGM was released by Life Technologies at the end of 2010.
•In case of PGM by Life Technologies Inc. change in PH is detected by the semiconductor
installed in each well of the chip.
•Change in PH occurs by the release of the H+ during the addition of nucleotide to the template.
Addition of the nucleotide is quantified (specific change in voltage corresponding to the PH
change by release of 1 proton) and therefore multiple addition of same nucleotide can be
detected with accuracy[1][7].
•Each time the chip was flooded with one nucleotide after another, if it is not the correct
nucleotide, no voltage will be found. Also voltage change corresponding to each type of
nucleotide in different[1][7].
•Library preparation involve amplification by emulsion PCR and addition of adaptors and
barcodes for identification of sample during multiplexing.](https://image.slidesharecdn.com/nextgenerationsequencing-170219062723/75/Next-generation-sequencing-25-2048.jpg)
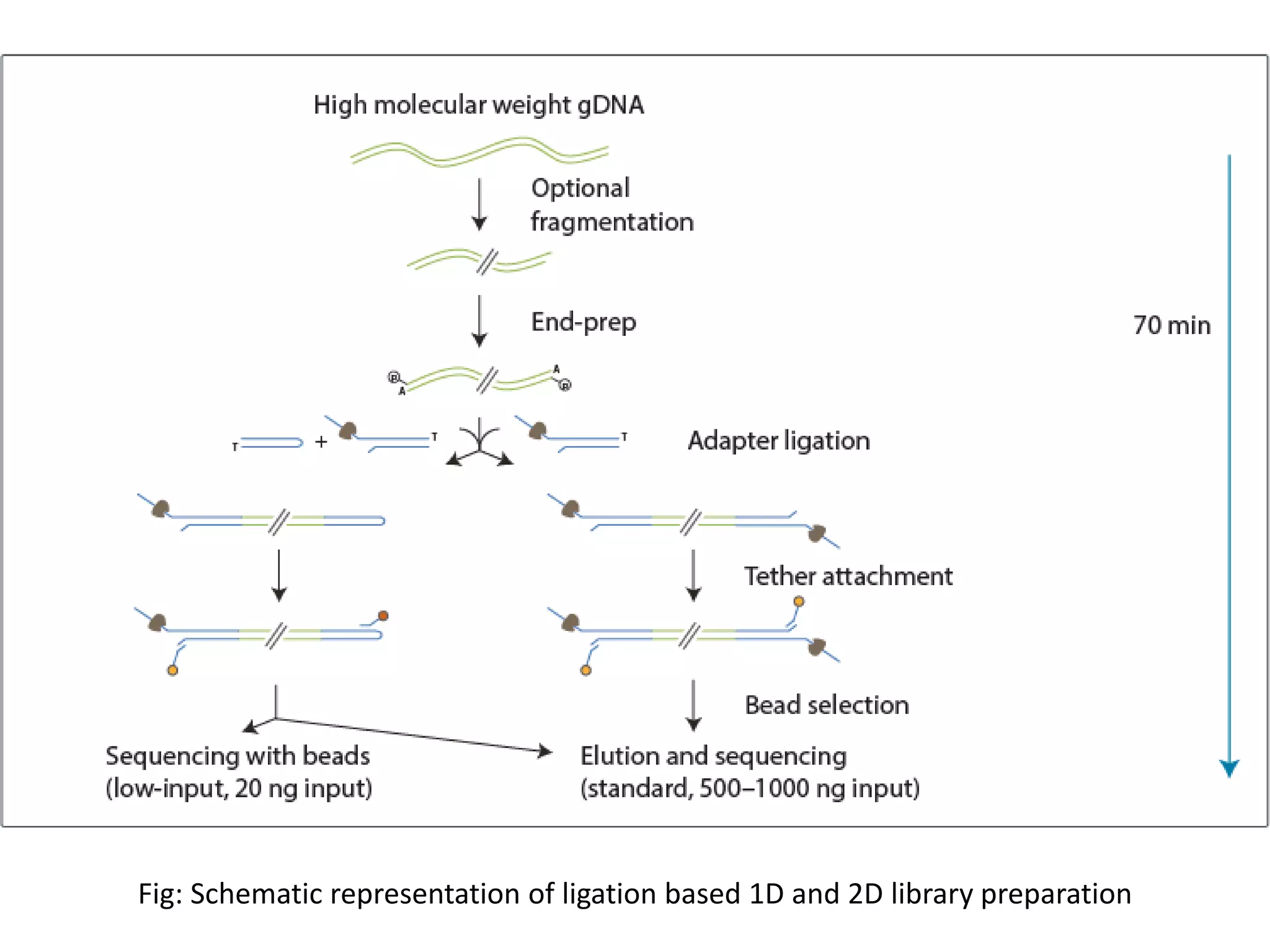
![•Smallest Sequencer ever
•Based on difference in the resistance to conduction of electrical current by the 4 types of bases
while passing through the Nanopore in single stranded form.
•DNA strand is pulled through the nanopore by the enzyme, one base at a time.
•In some libraries DNA strands are bound by hairpin loop so that both the strands are read in a
single go thereby increasing the accuracy and efficiency of sequencing[8].](https://image.slidesharecdn.com/nextgenerationsequencing-170219062723/75/Next-generation-sequencing-29-2048.jpg)
![Fig: Diagrammatic representation of structure of Nanopore and its attachment in the membrane[9].
Fig: Diagrammatic representation showing effect of small molecule on the electrical conductivity of nanopore[9]
Fig: Diagrammatic representation showing effect of larger molecule on the electrical conductivity of nanopore[9].](https://image.slidesharecdn.com/nextgenerationsequencing-170219062723/75/Next-generation-sequencing-31-2048.jpg)
![Fig: Passing of DNA strand through the nanopore causes electrical disturbances in the pore
which is characteristic for each type of base and is recorded for base calling[9].](https://image.slidesharecdn.com/nextgenerationsequencing-170219062723/75/Next-generation-sequencing-32-2048.jpg)