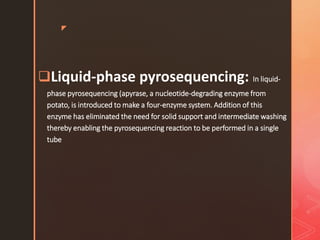
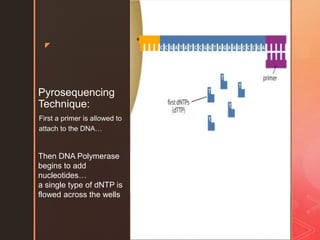
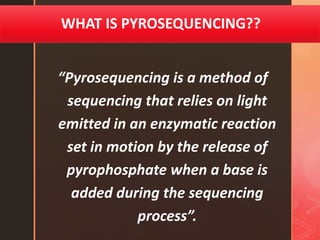
Pyrosequencing is a sequencing method that detects light signals from enzymatic reactions triggered by nucleotide additions during DNA synthesis. It was developed in 1996 and allows high-throughput sequencing. There are solid and liquid phase variants, with the latter using an additional enzyme to eliminate washing steps. The process involves preparing DNA fragments, attaching to beads, amplification by PCR, and sequencing by flowing nucleotides over wells containing DNA-coated beads and enzymes, detecting light signals with each nucleotide incorporation.

![z
HISTORY
The technique was developed by Mostafa Ronaghi and Pål Nyrén at
the Royal Institute of Technology in Stockholm in 1996.
A second solution-based method for Pyrosequencing was described
in 1998 (In this alternative method, an additional enzyme apyrase is
introduced.
A third microfluidic variant of the Pyrosequencing method was
described in 2005[5] by Jonathan Rothberg.
This allowed for high-throughput DNA sequencing and an
automated instrument was introduced to the market. This became
the first next generation sequencing instrument starting a new era
in genomics research](https://image.slidesharecdn.com/pyrosequencing-180516141421/85/Pyrosequencing-3-320.jpg)