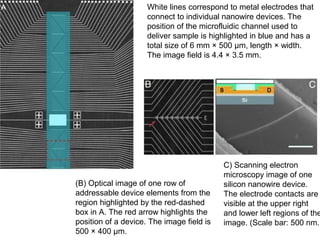
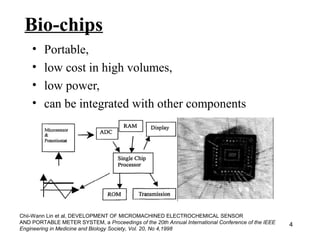
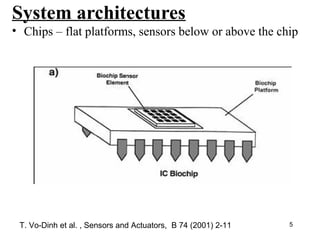
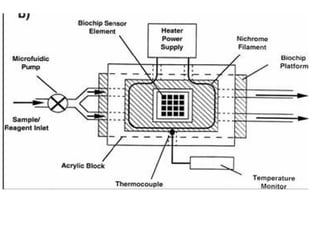
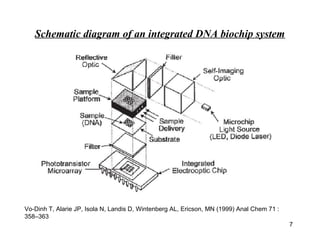
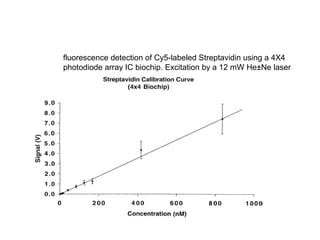
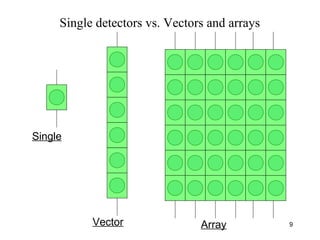
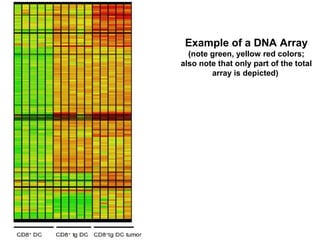
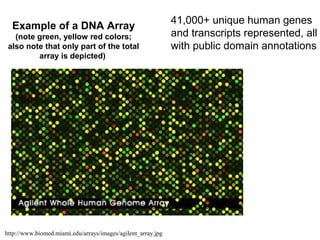
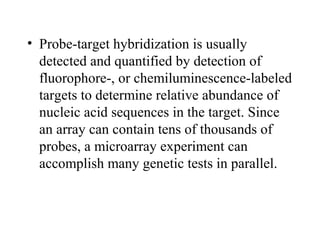
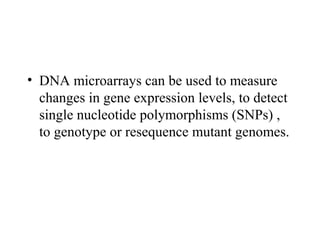
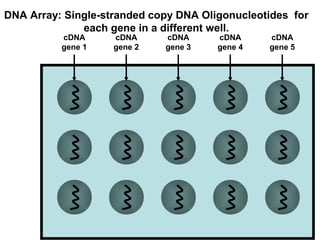
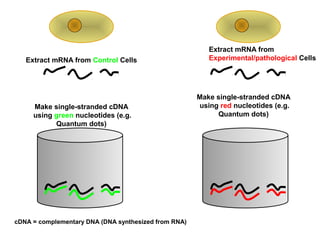
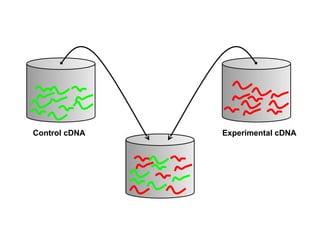
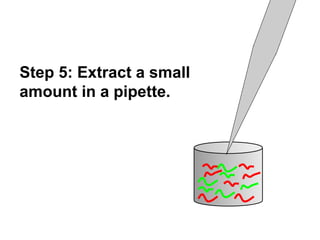
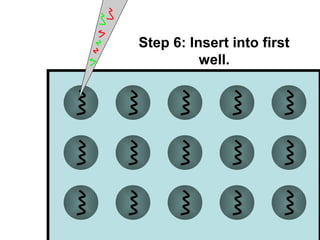
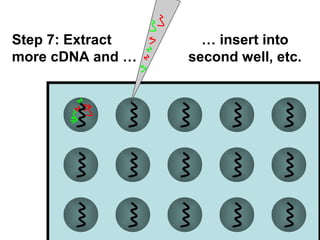
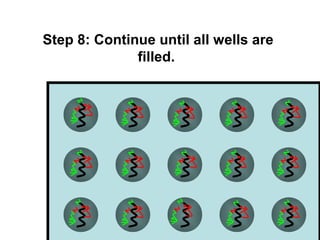
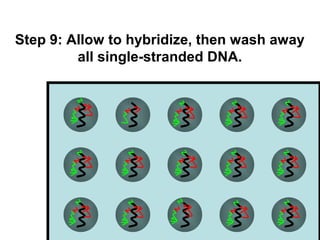
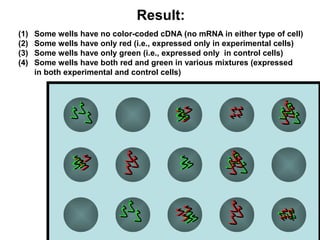
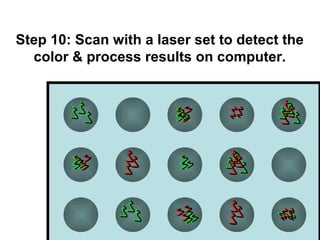
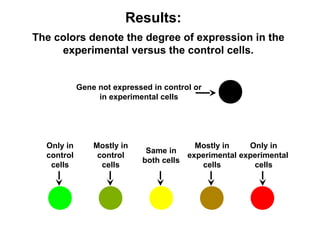
This document discusses bio-chips and DNA microarrays. It describes how DNA microarrays work by having thousands of probes attached to a surface to which cDNA from experimental and control samples can hybridize. The document outlines the steps of DNA microarray experiments including creating the array, extracting mRNA from samples, making cDNA, hybridizing the cDNA to the array, and scanning the array to assess gene expression differences between conditions. DNA microarrays allow analysis of expression levels of many genes in parallel.