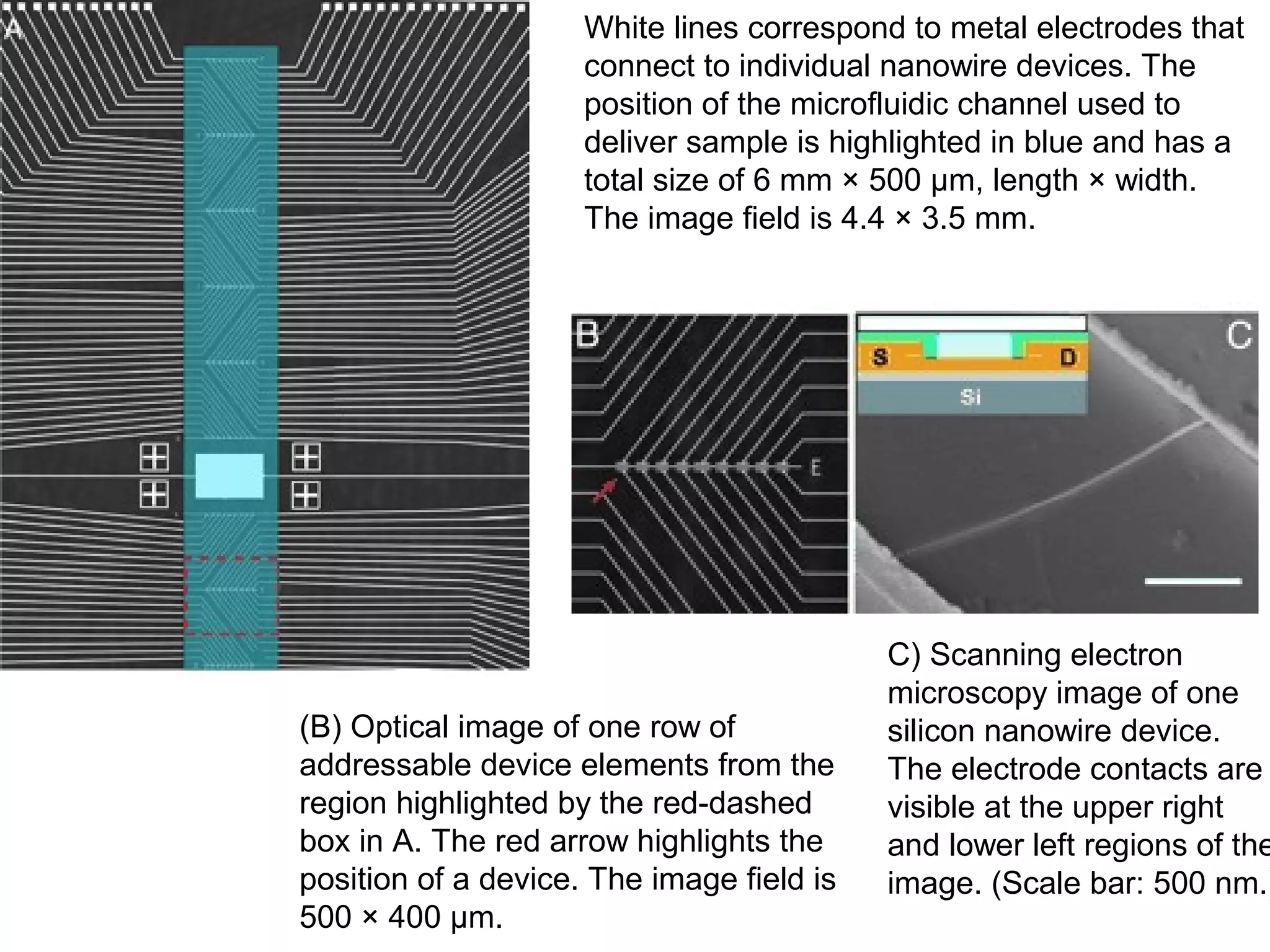
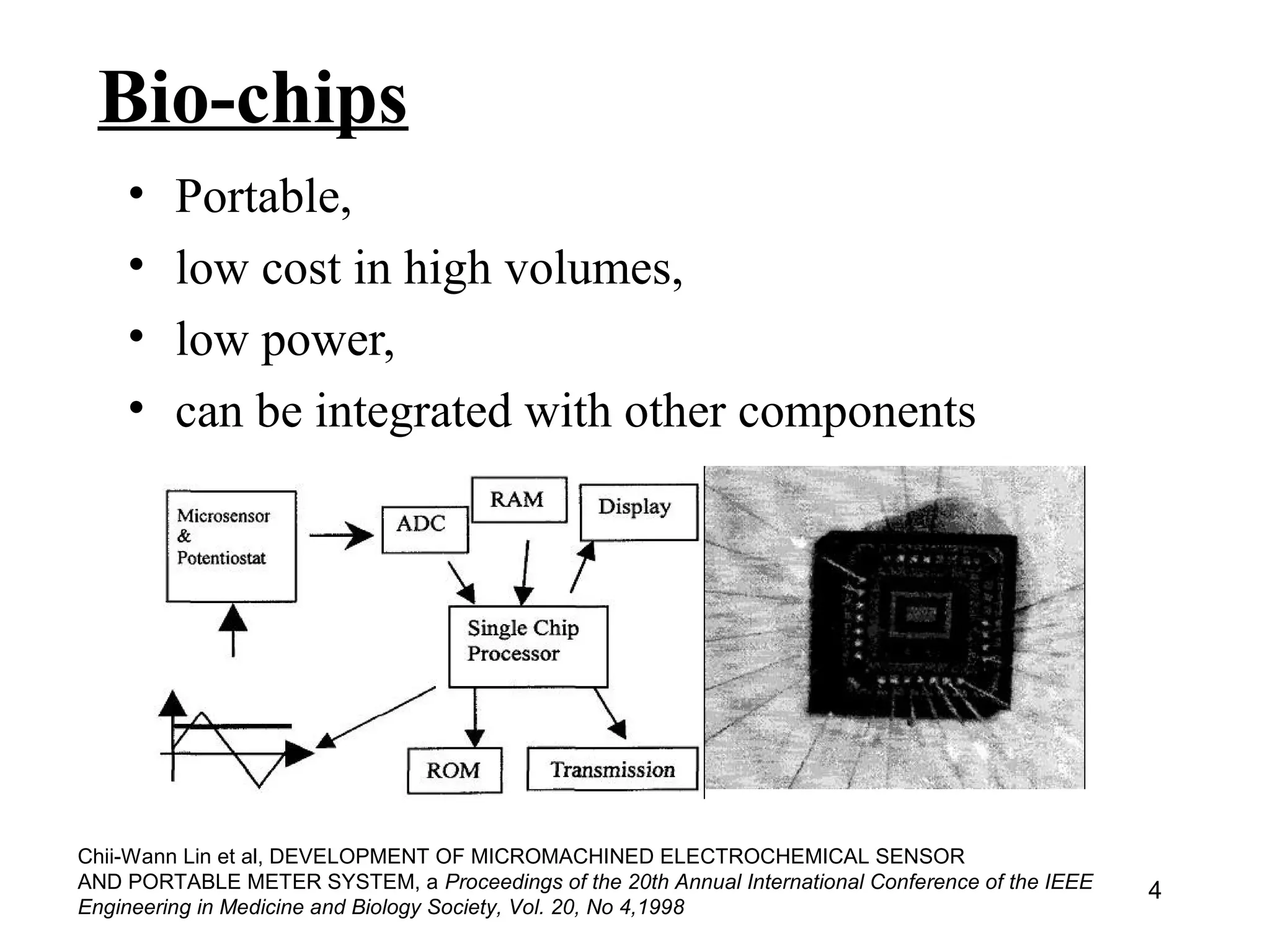
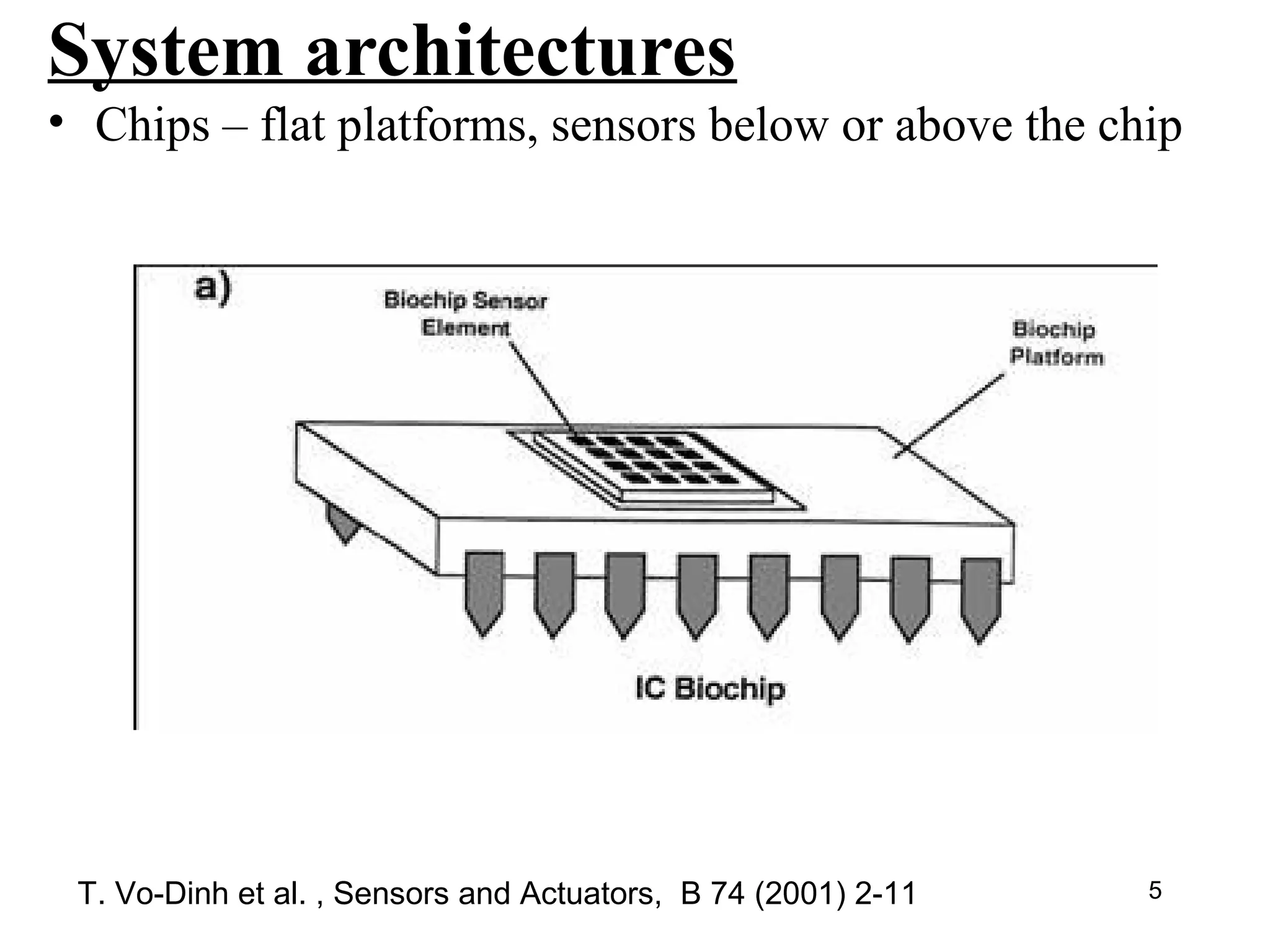
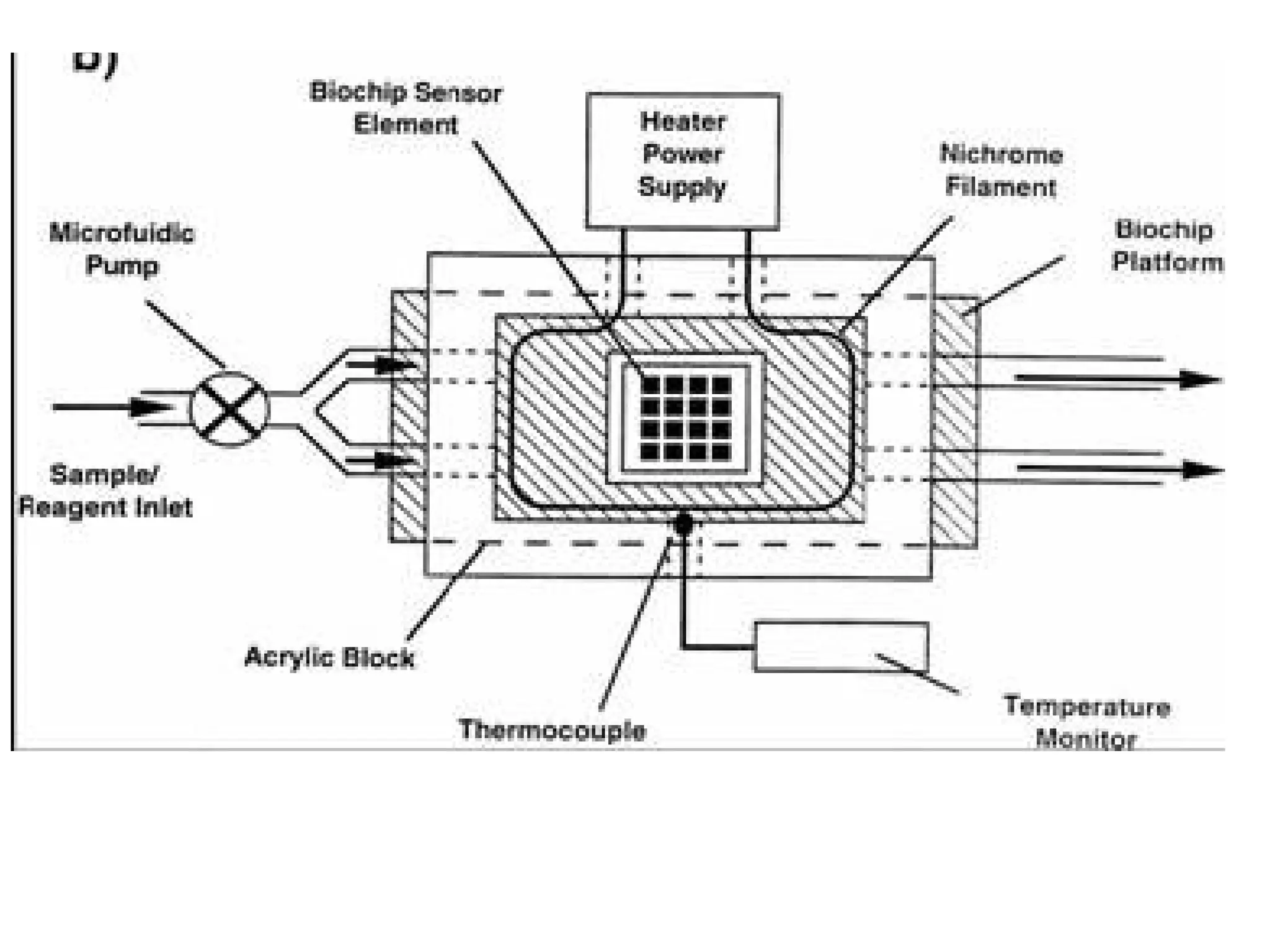
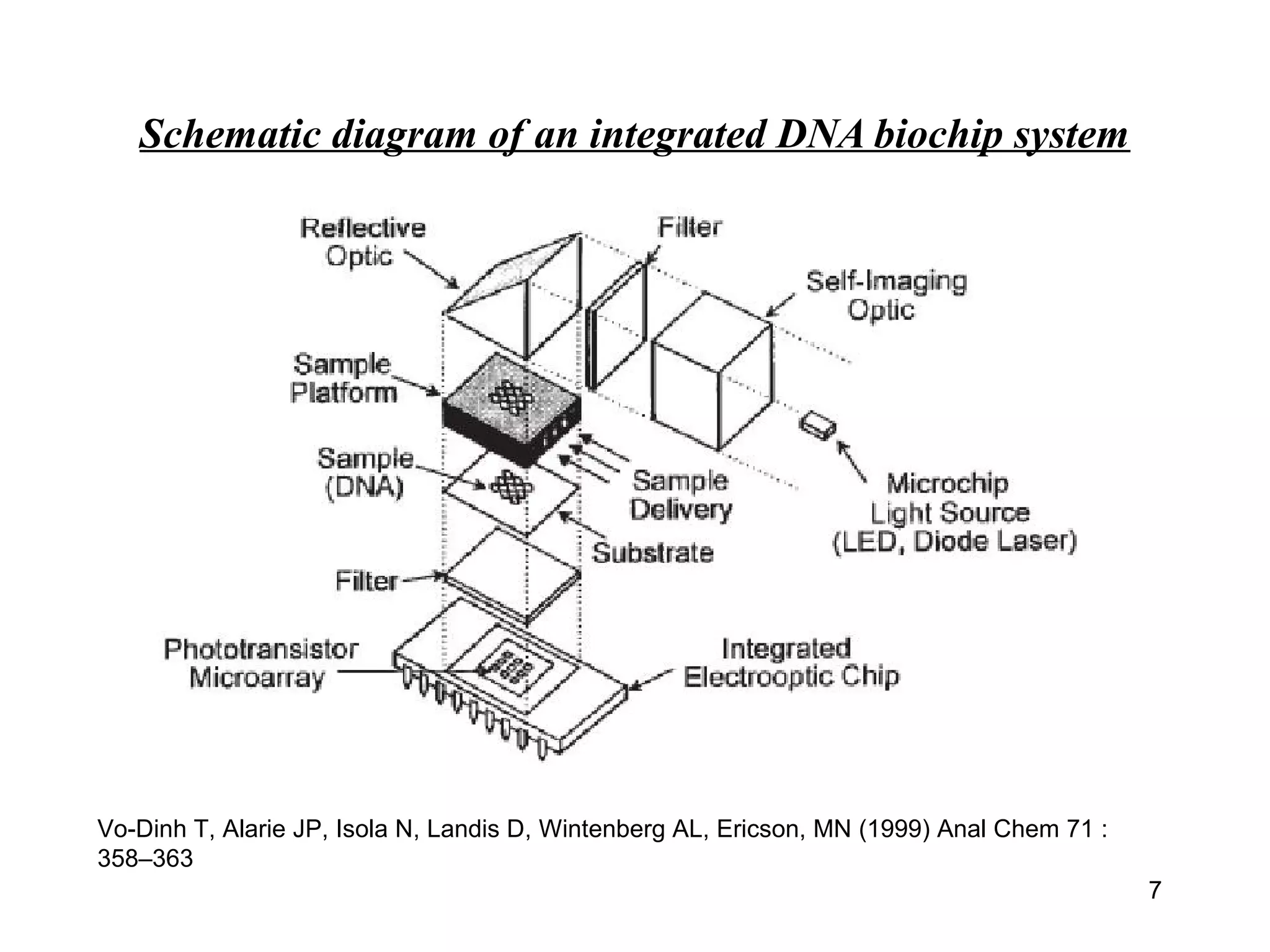
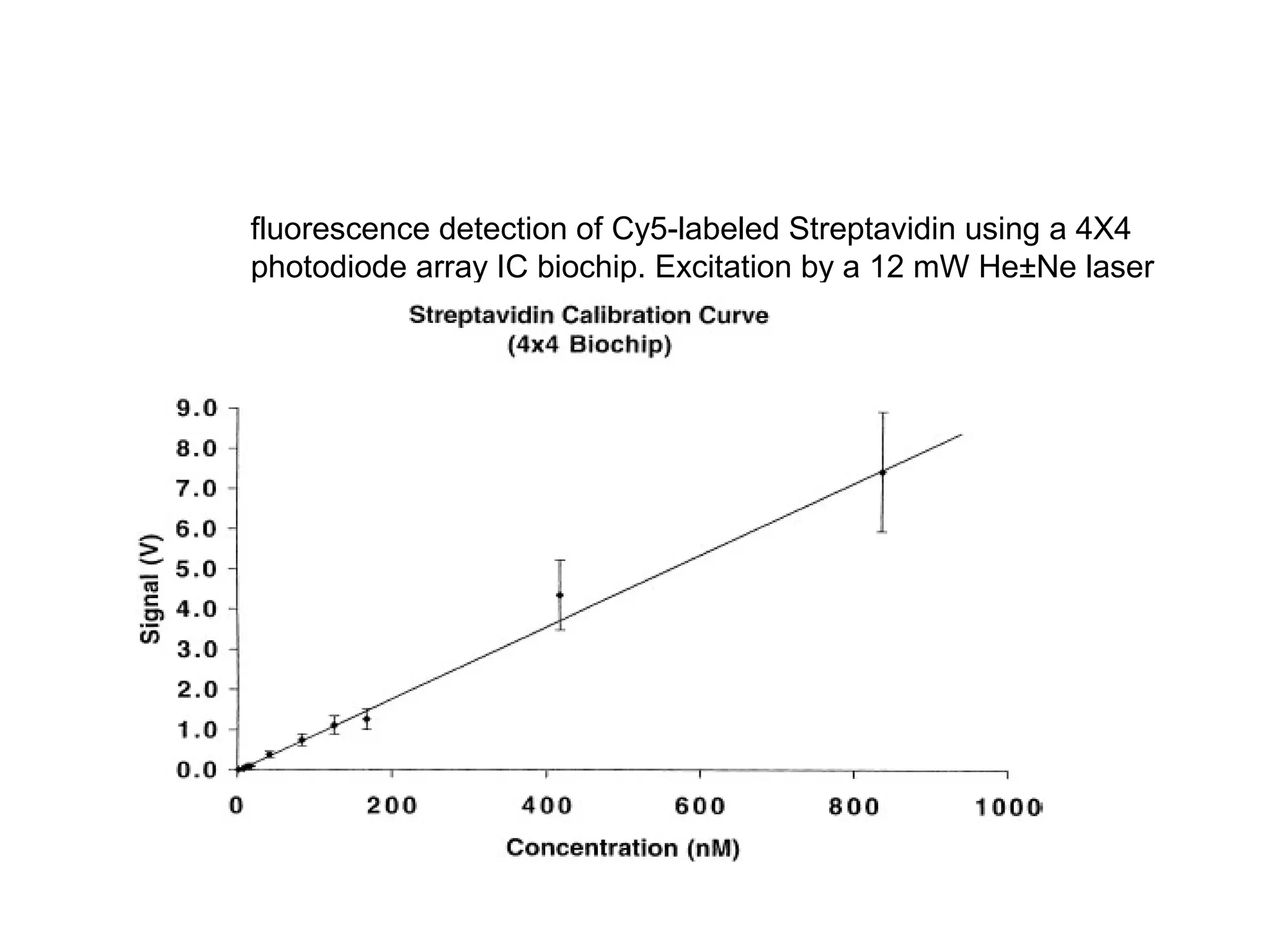
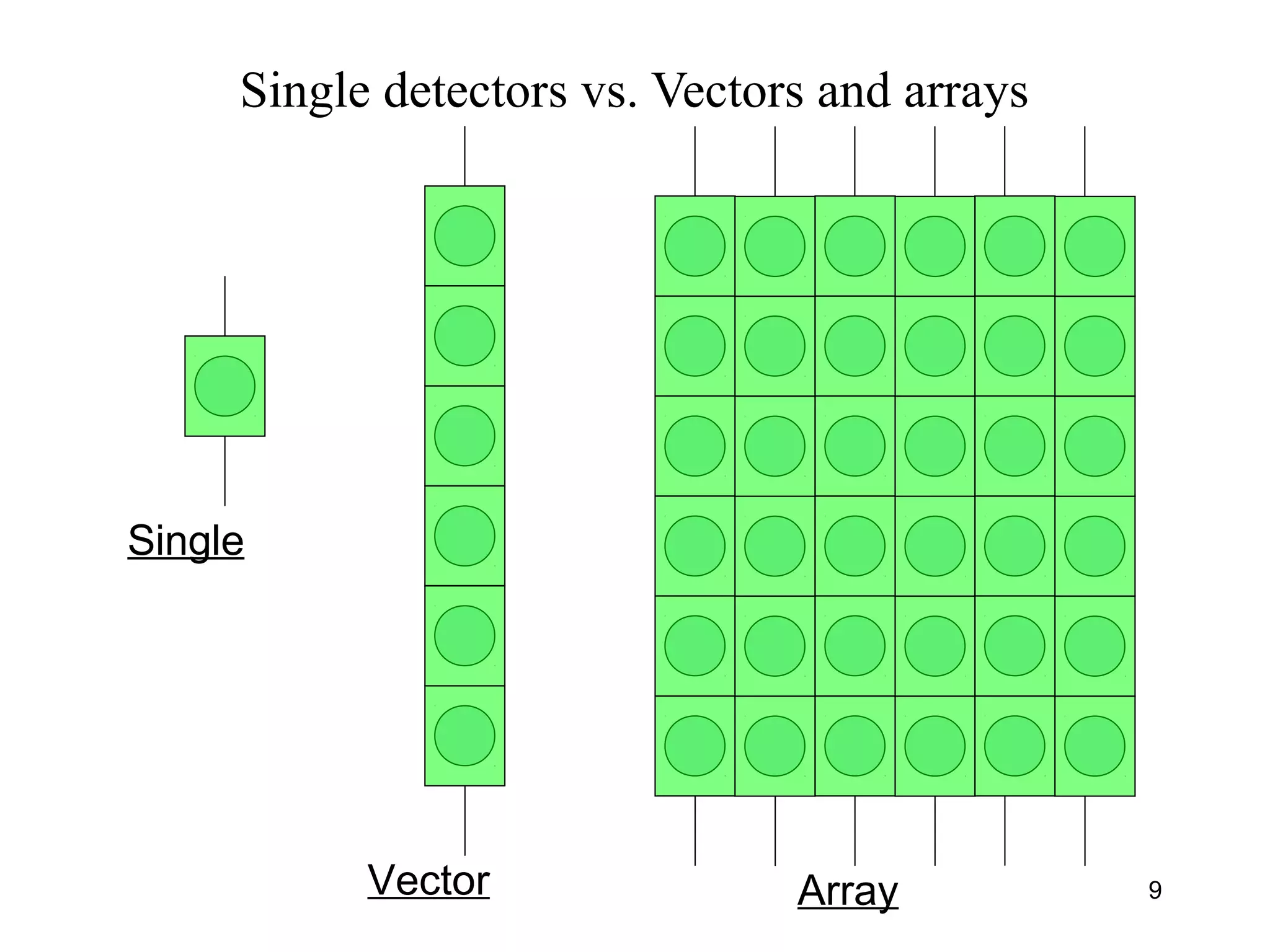
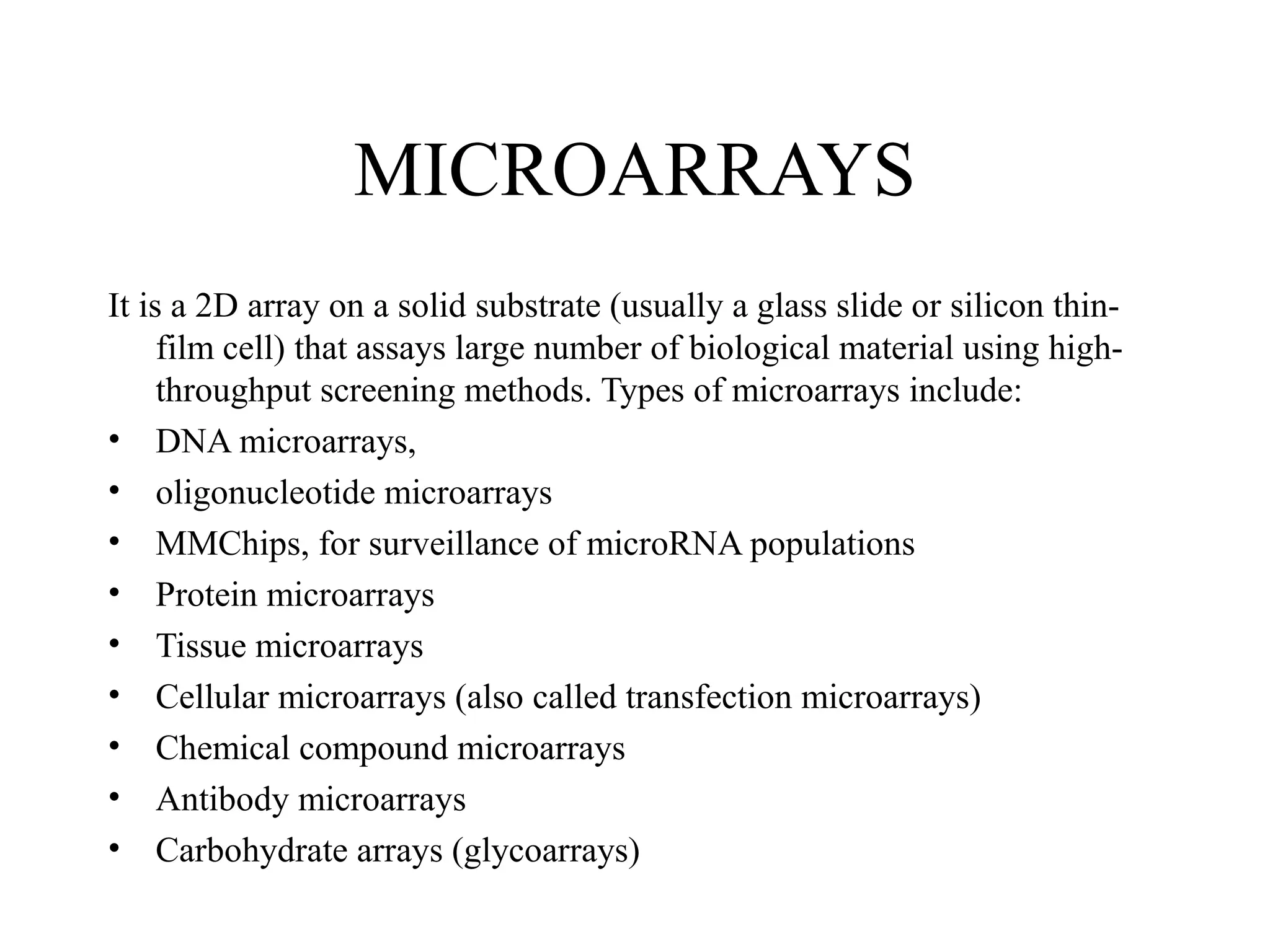
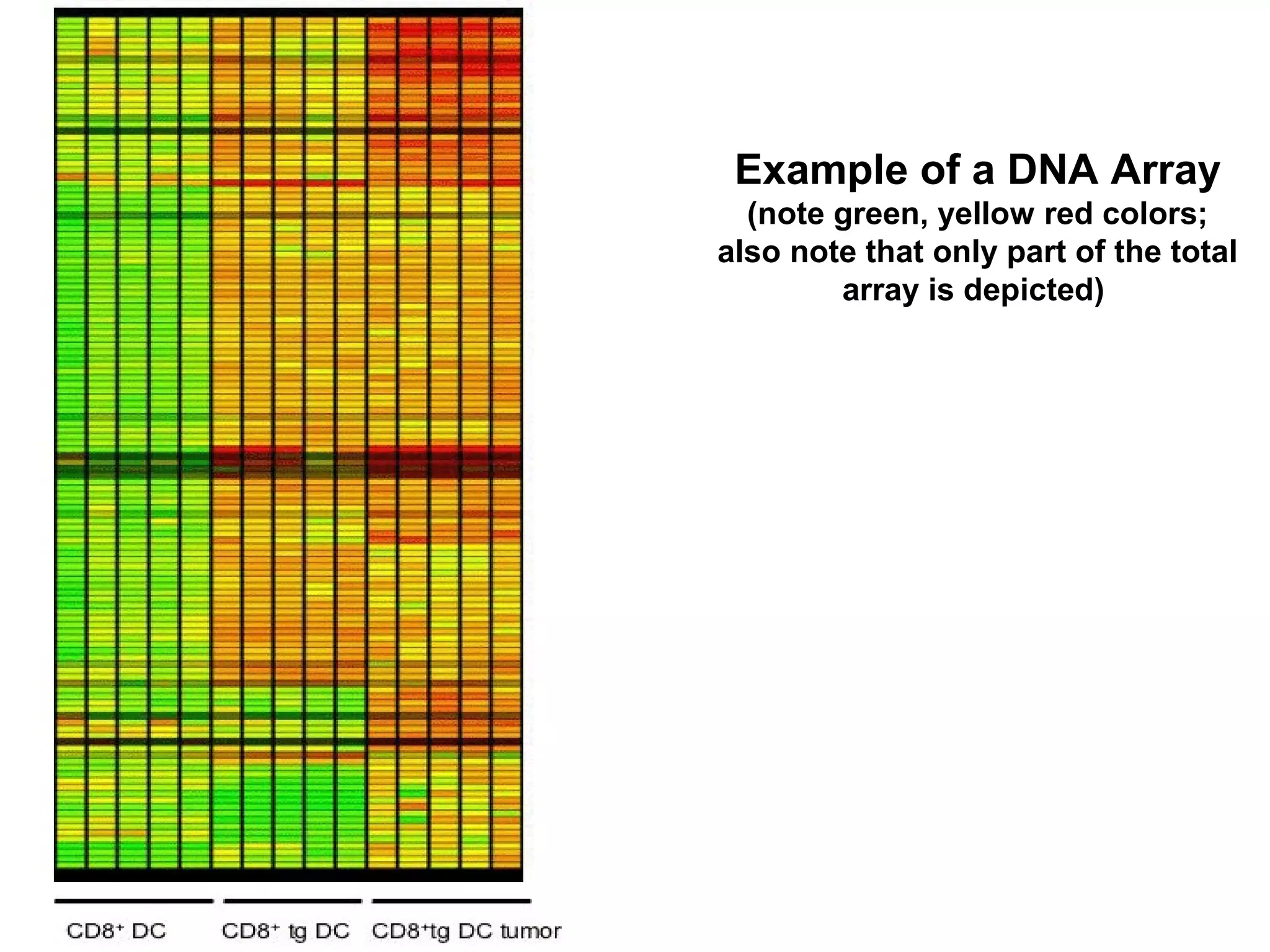
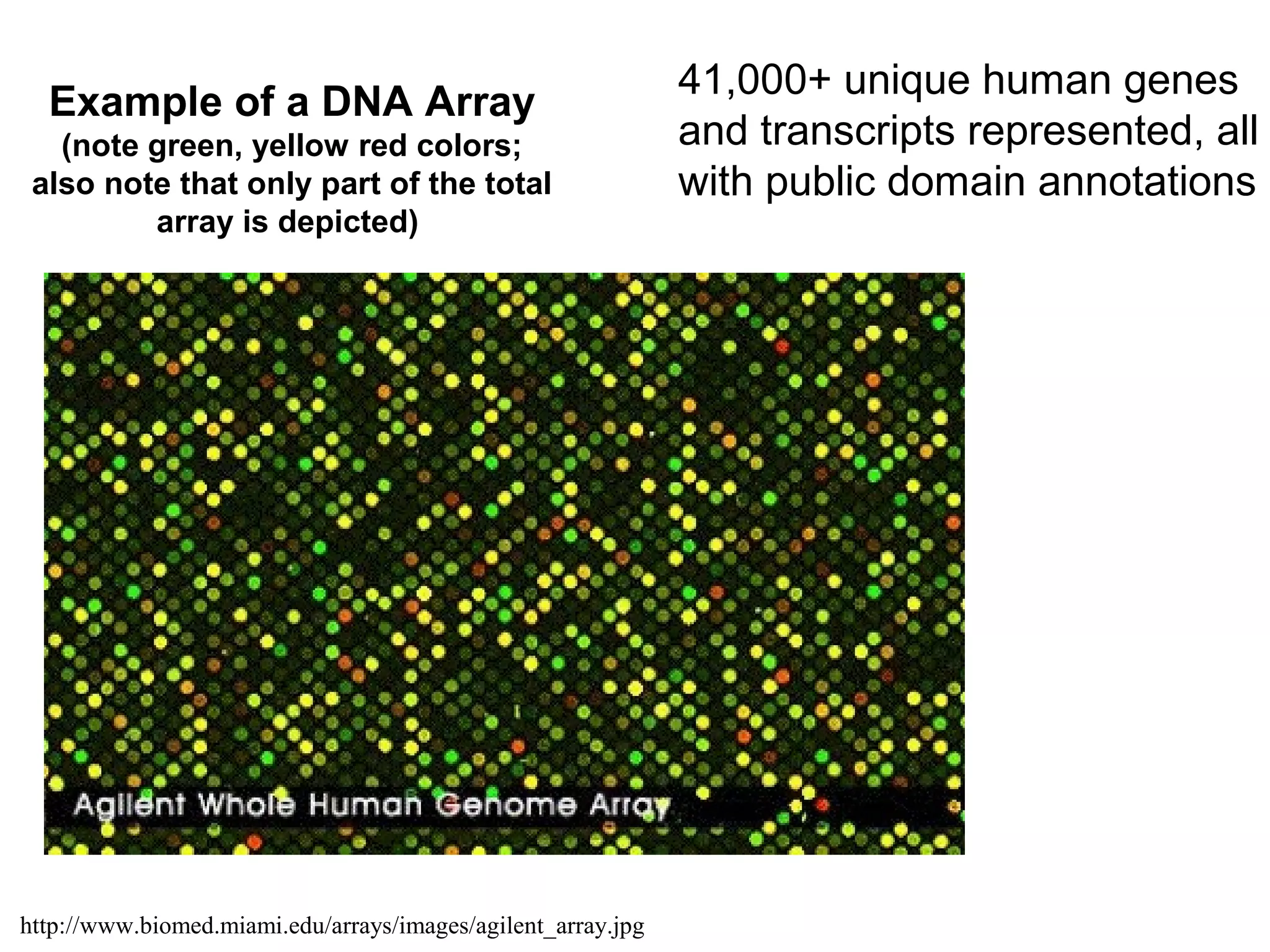
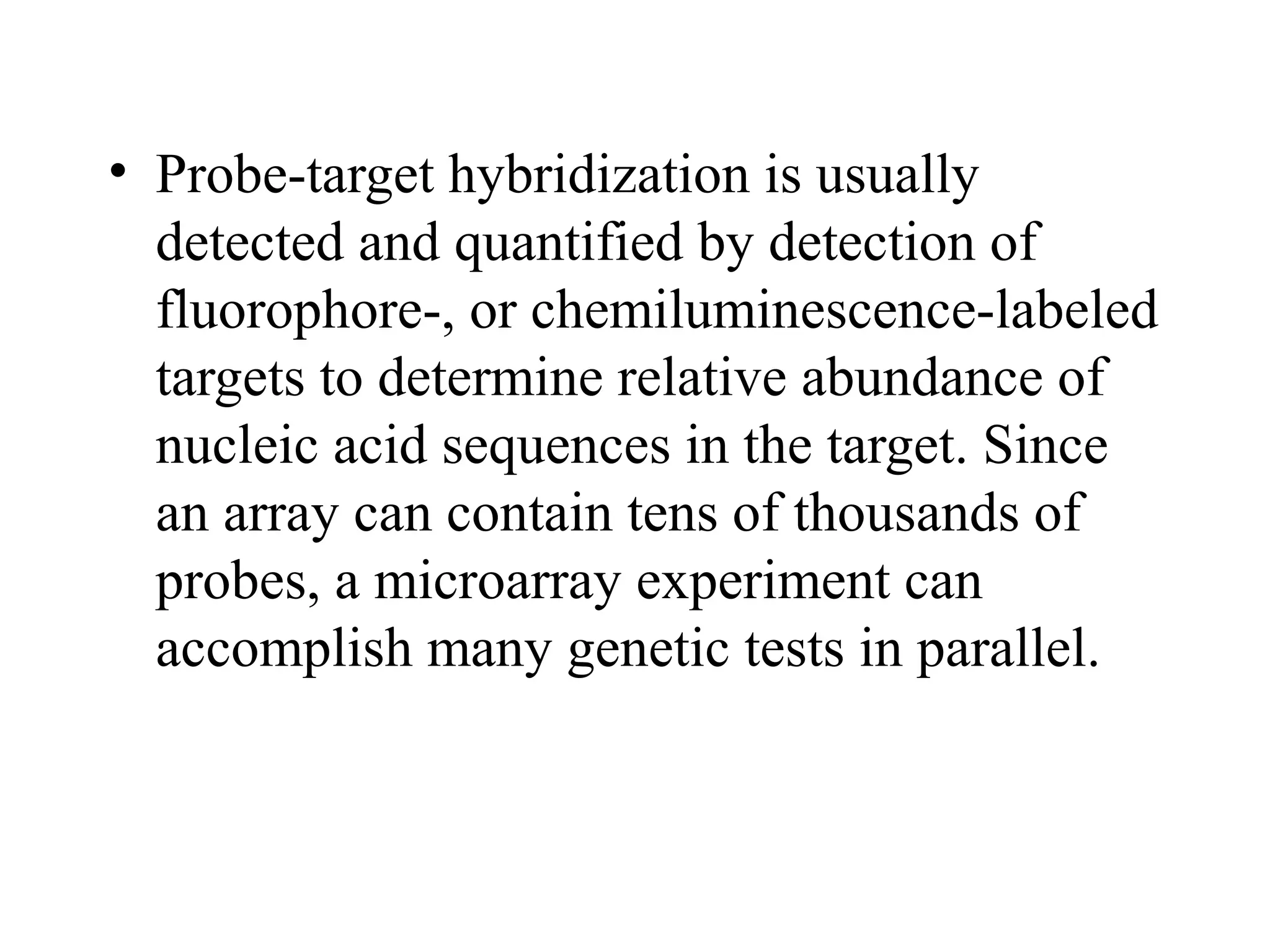
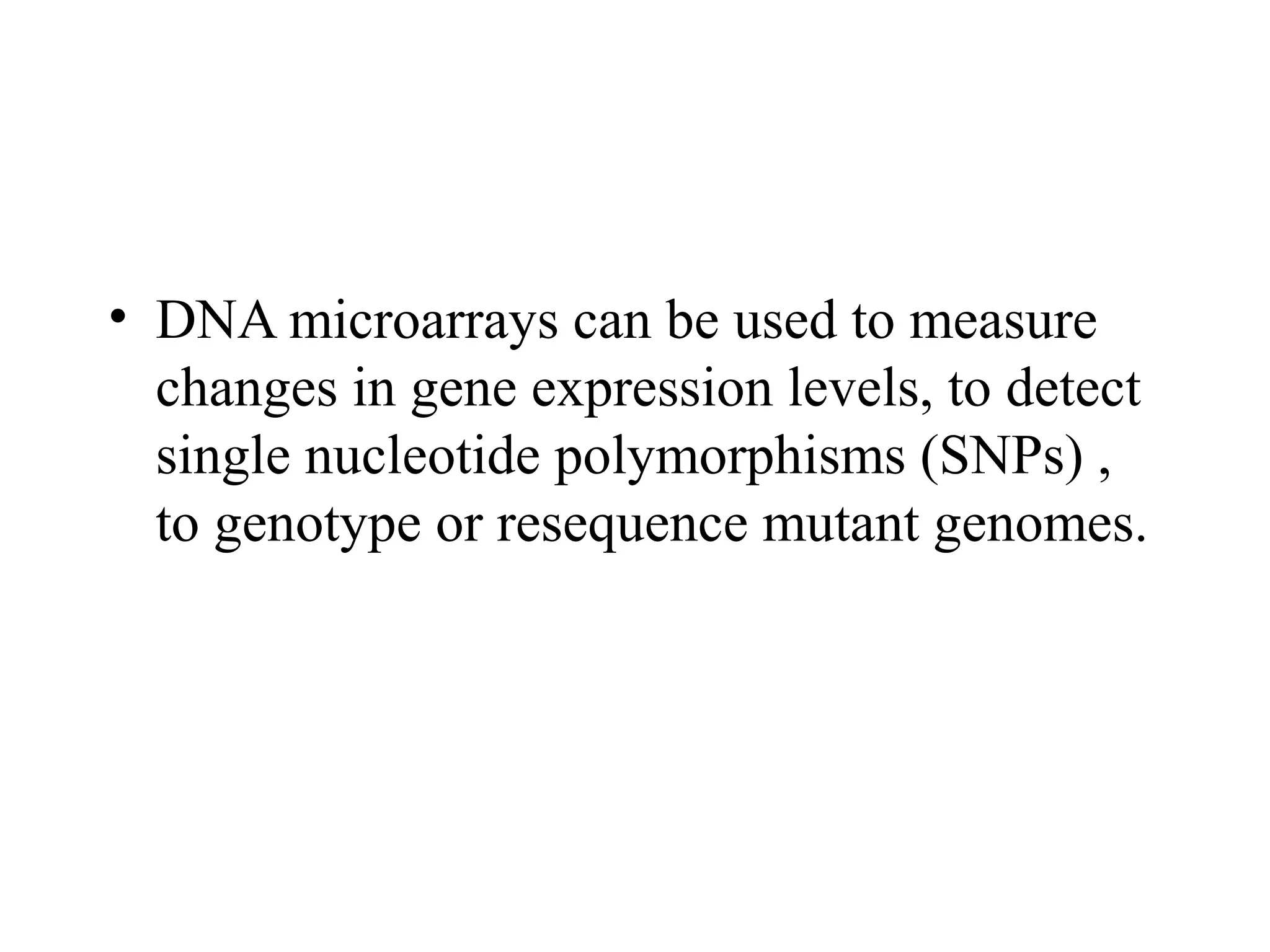
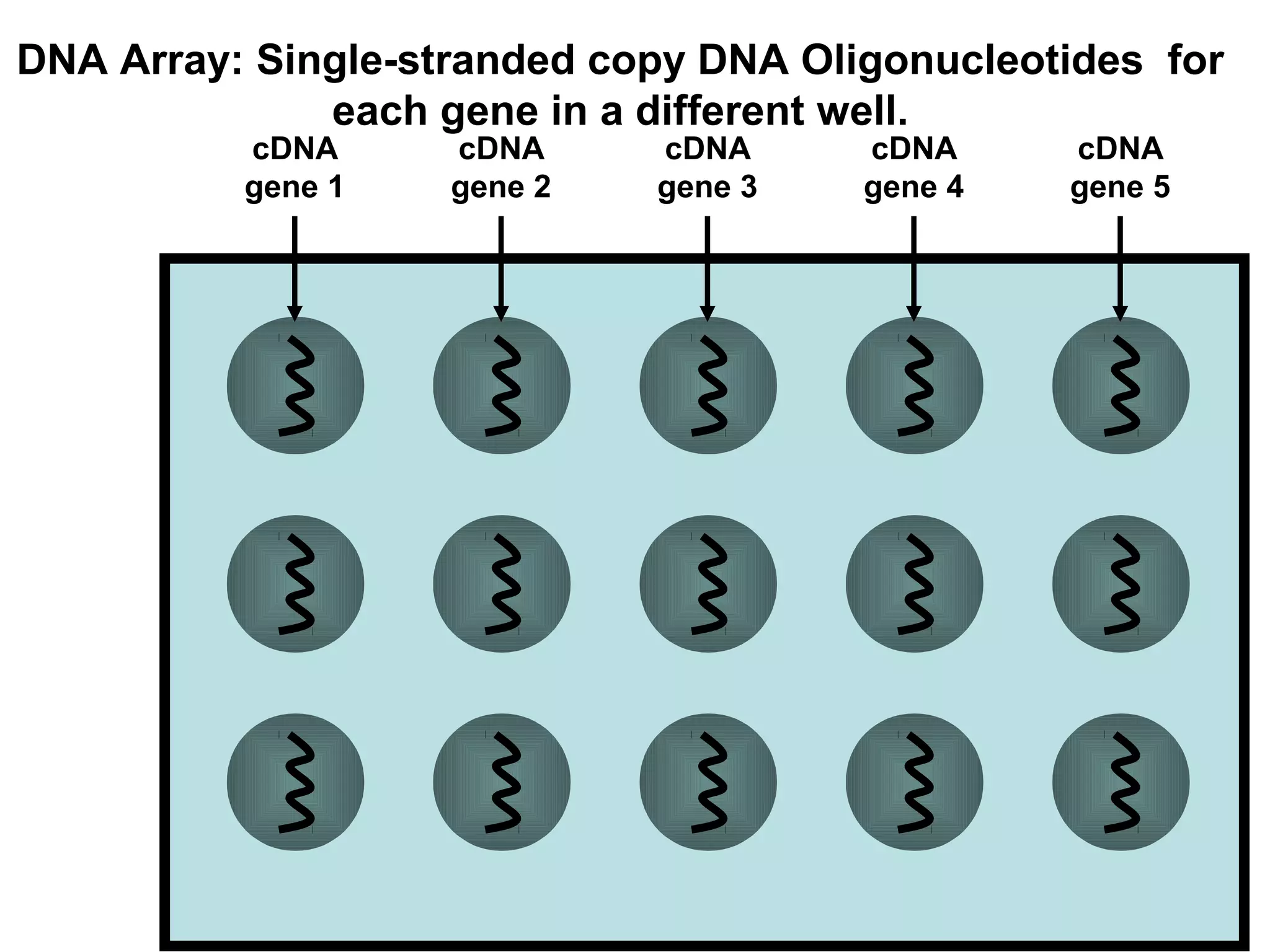
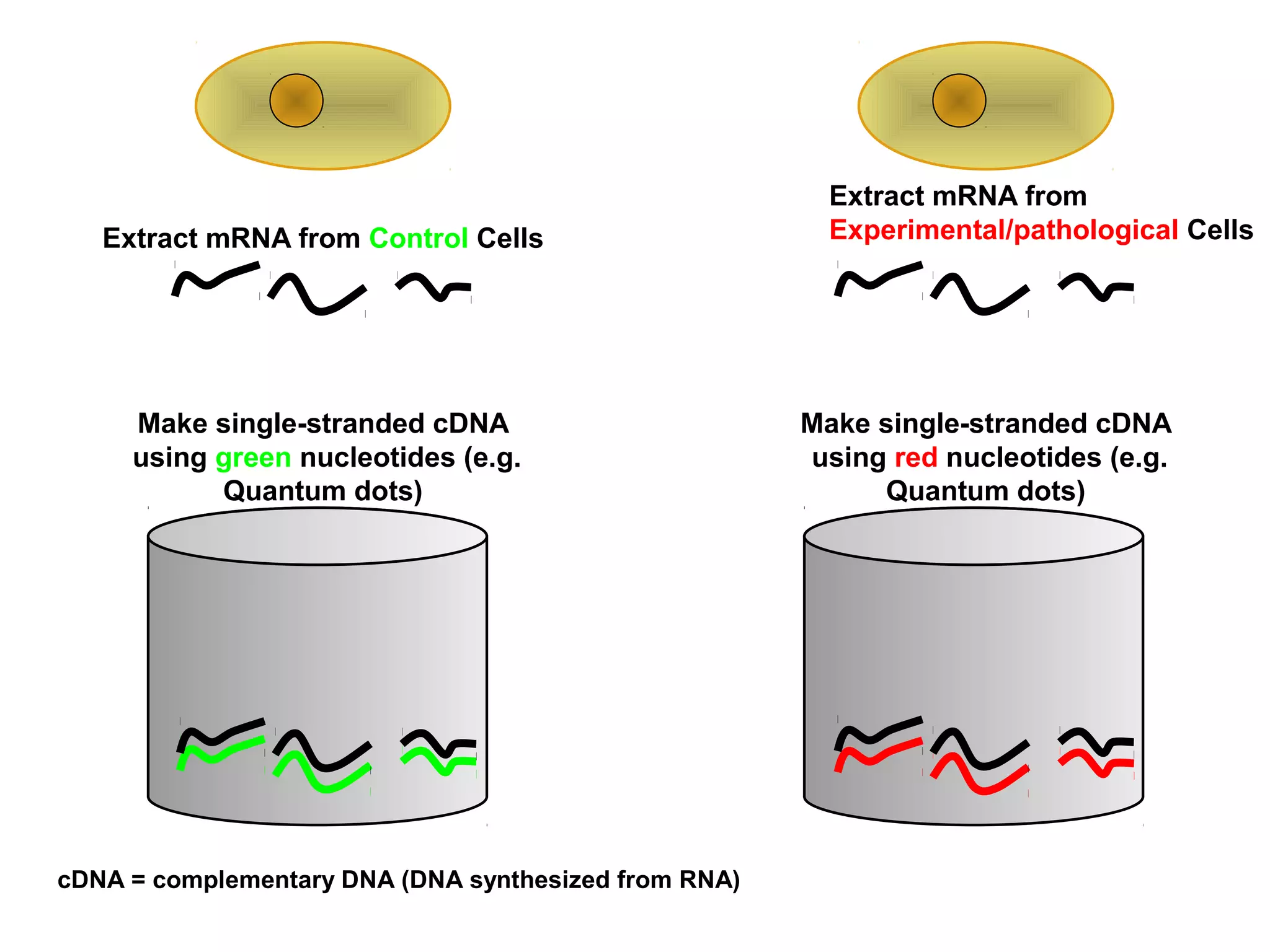
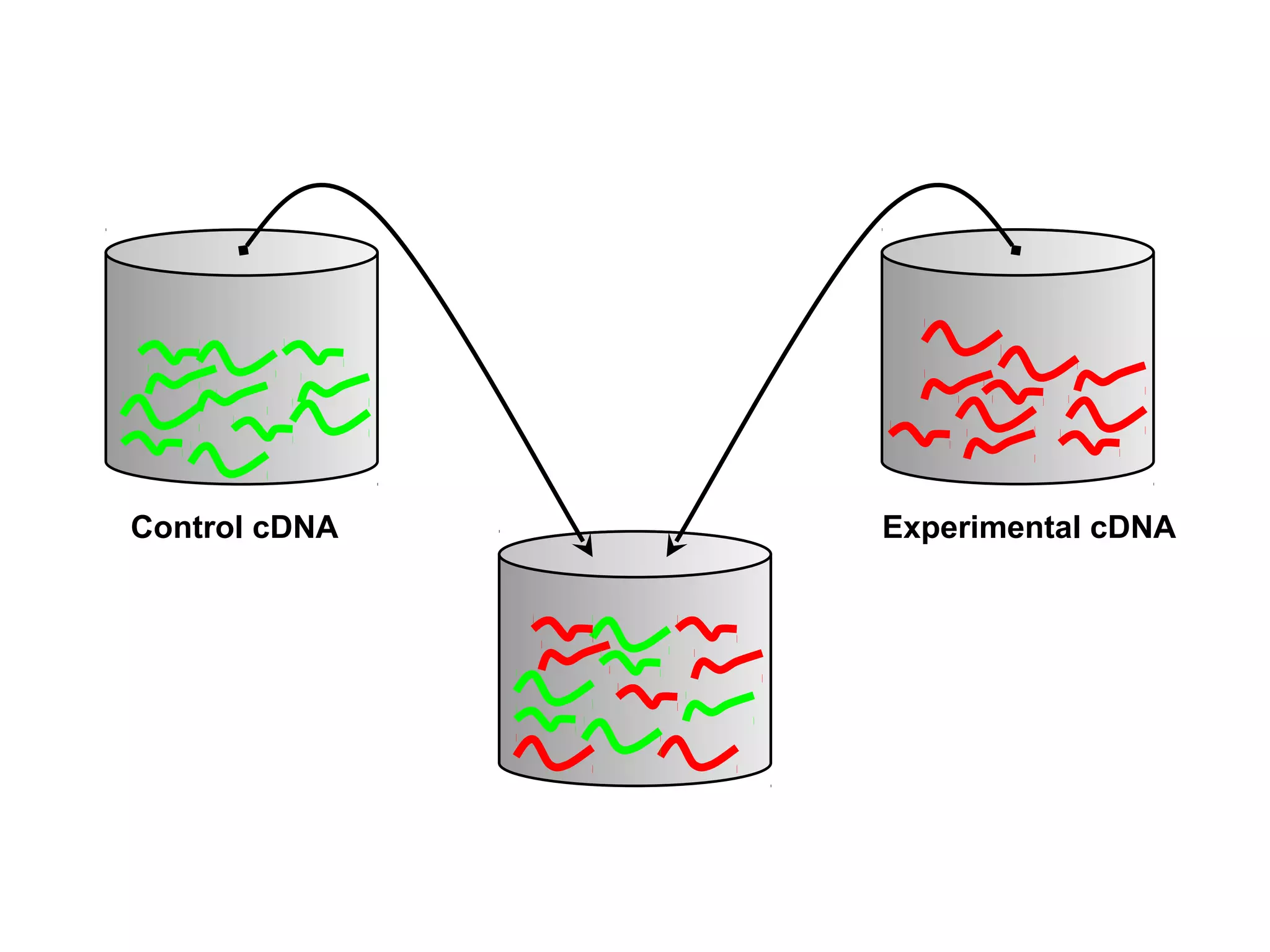
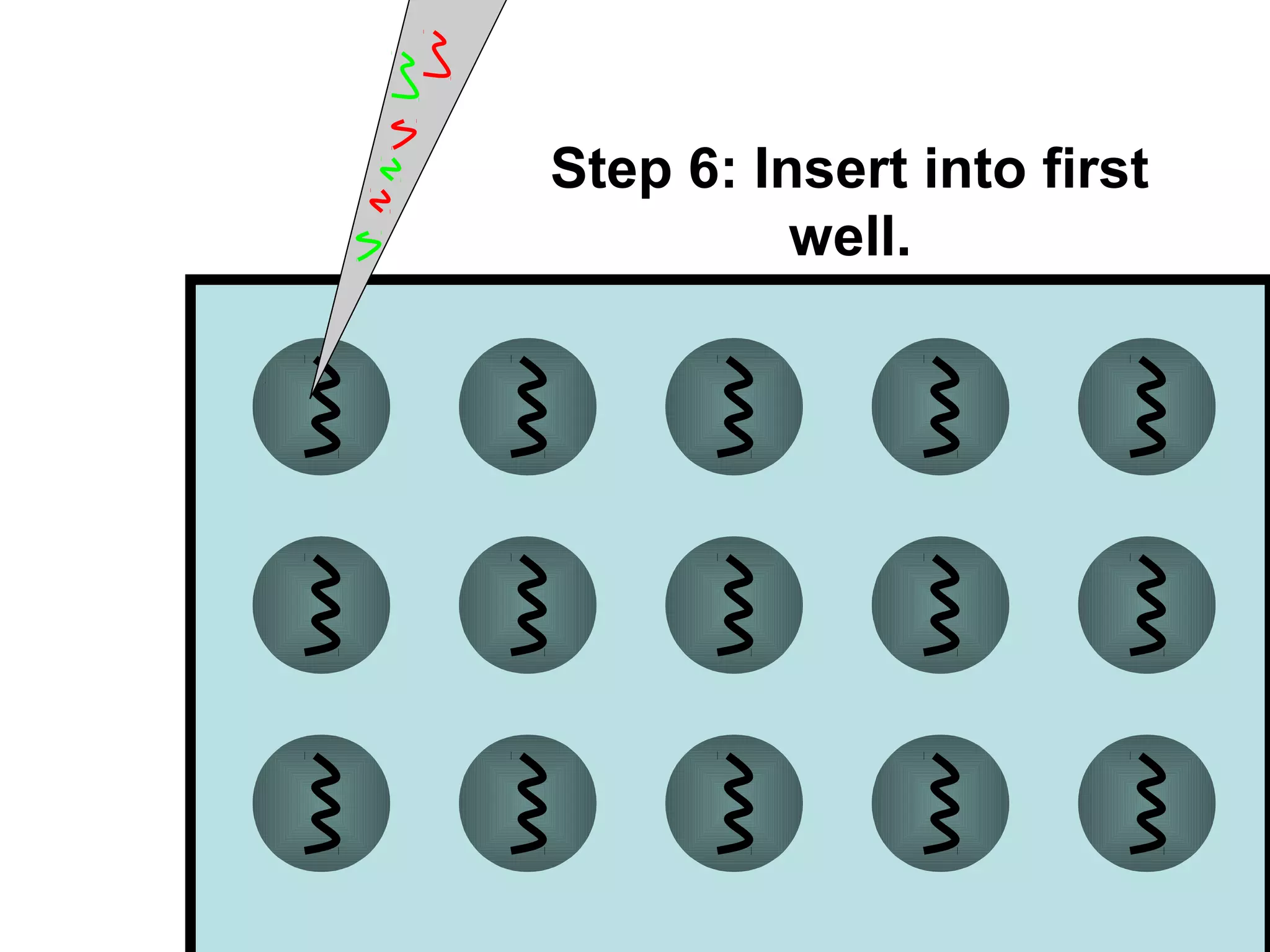
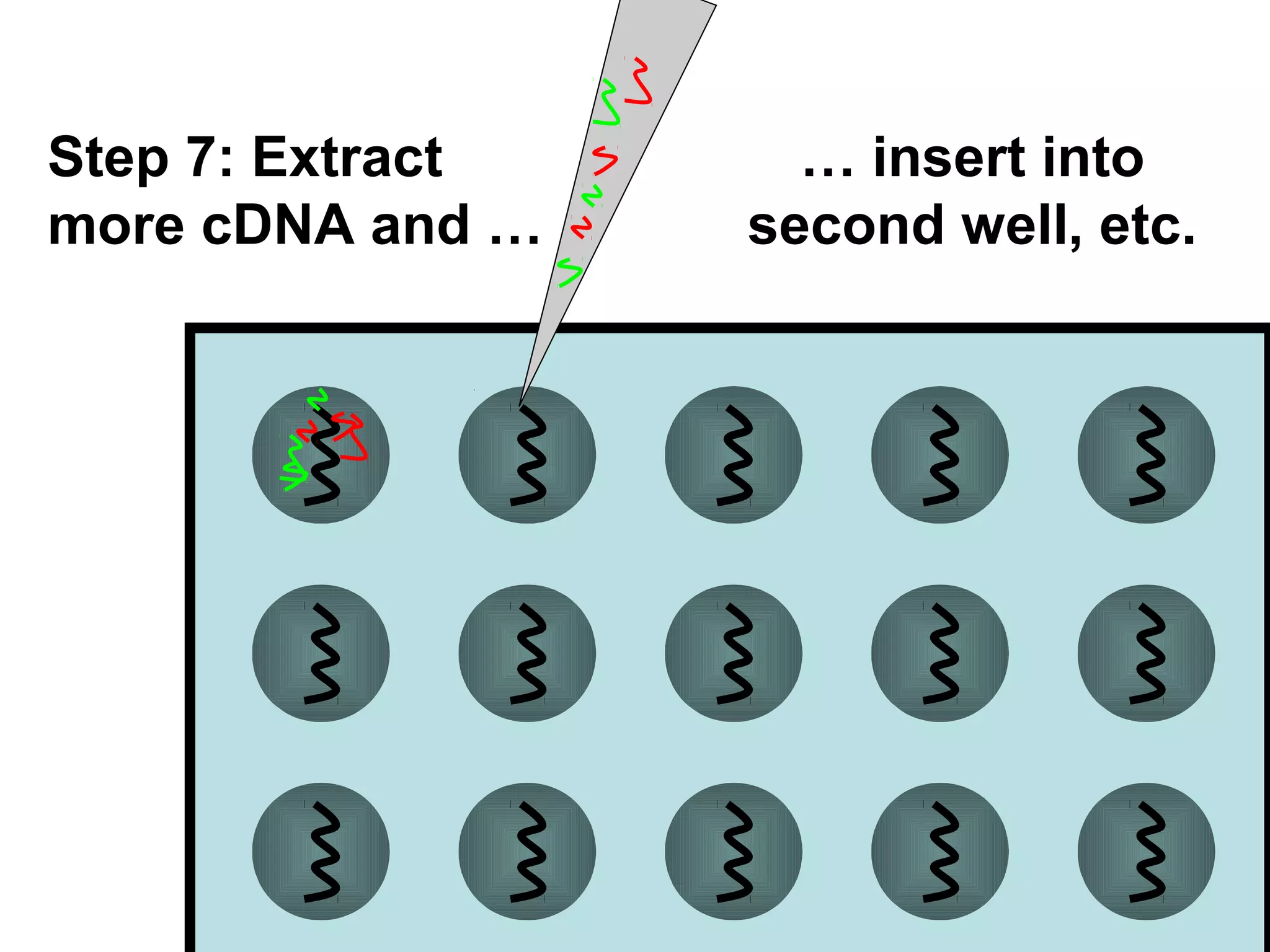
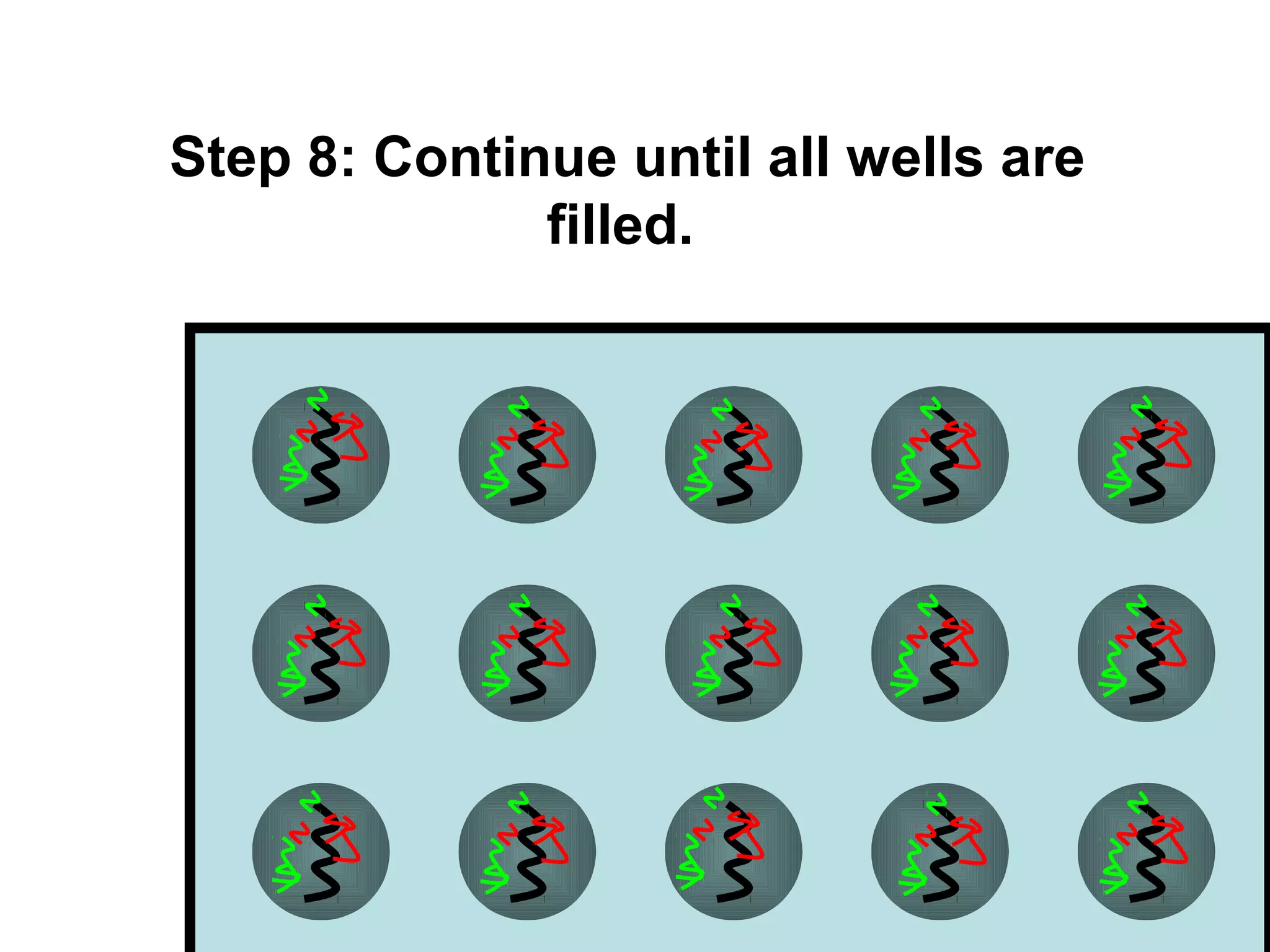
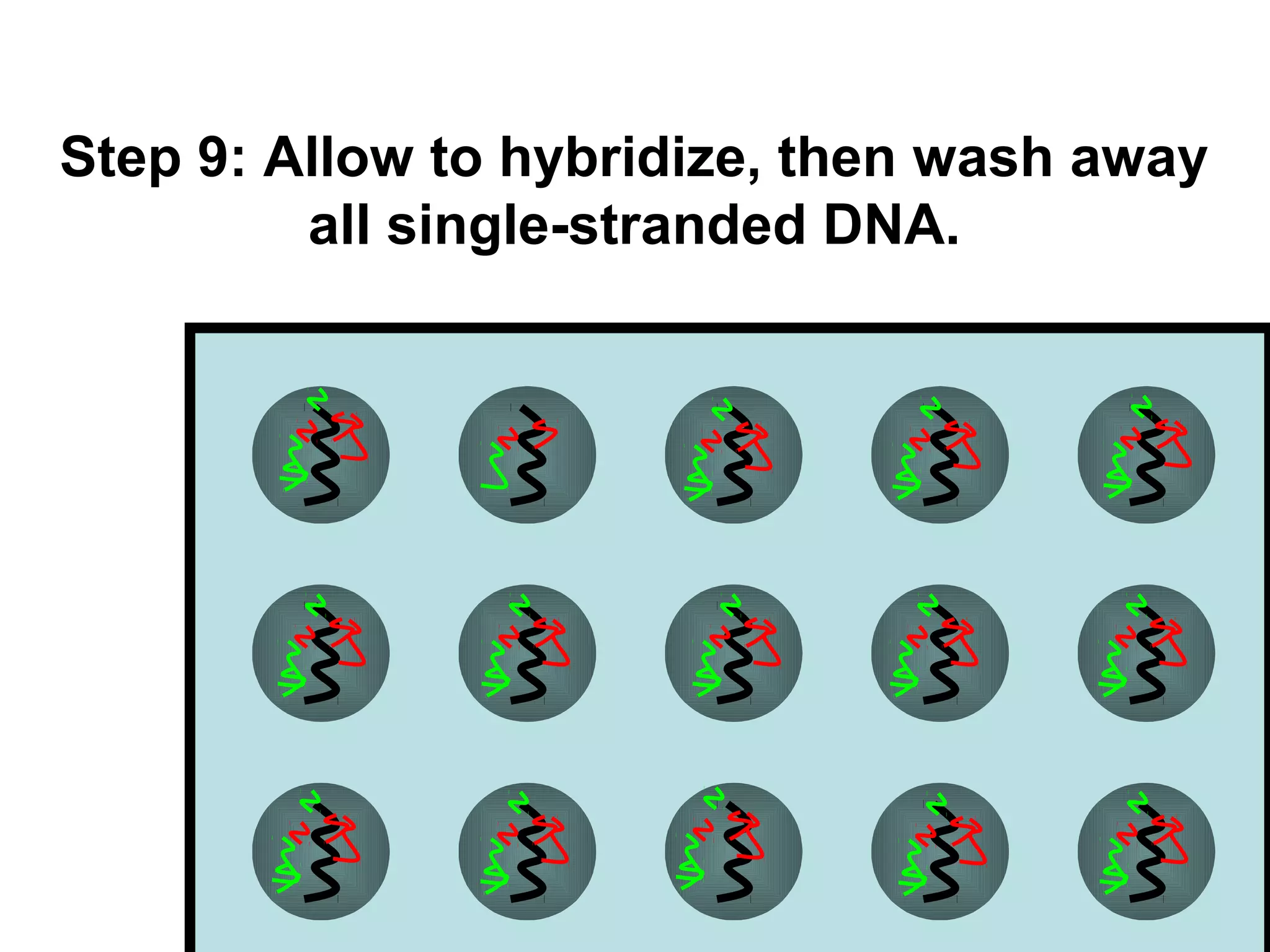
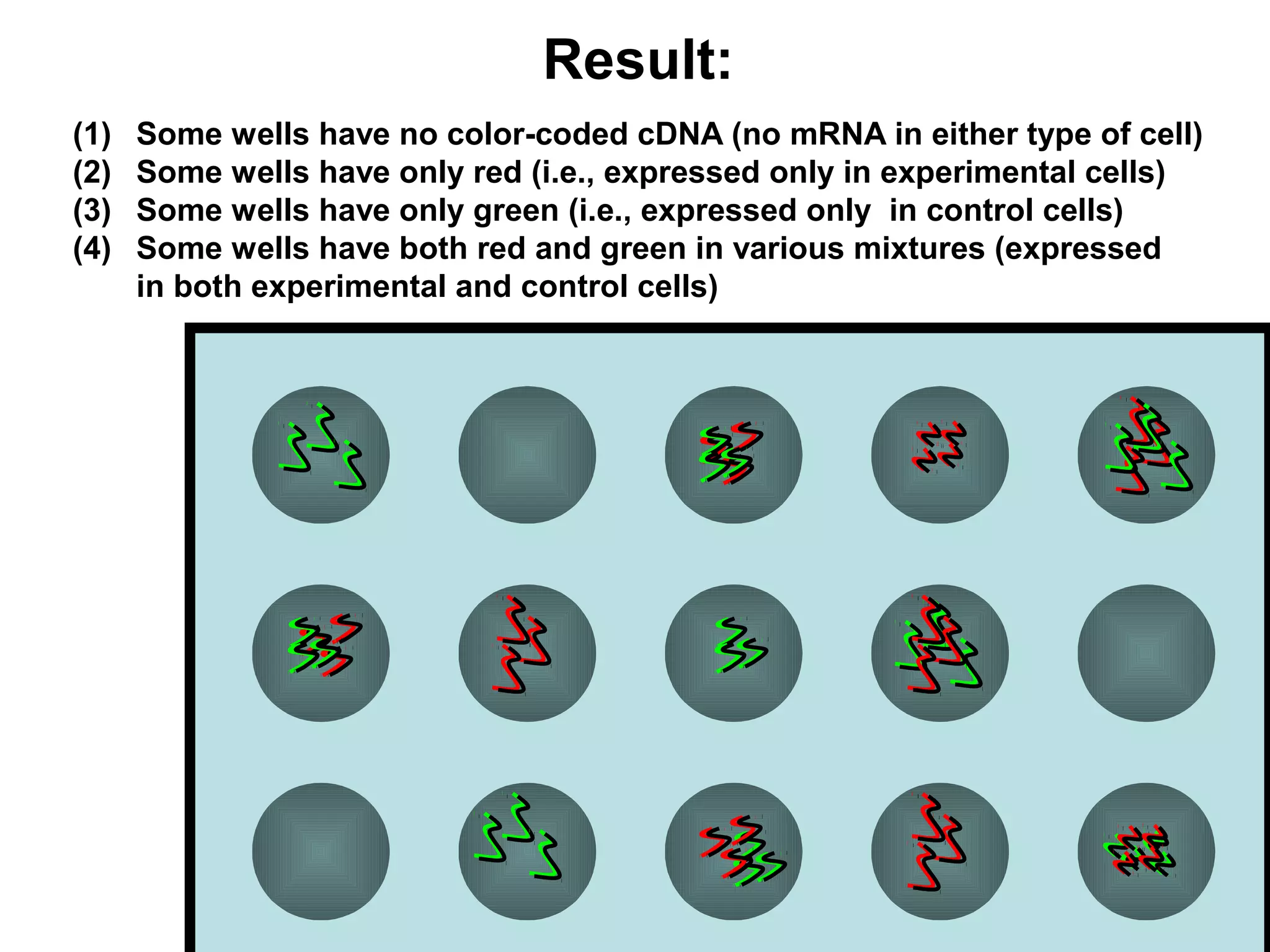
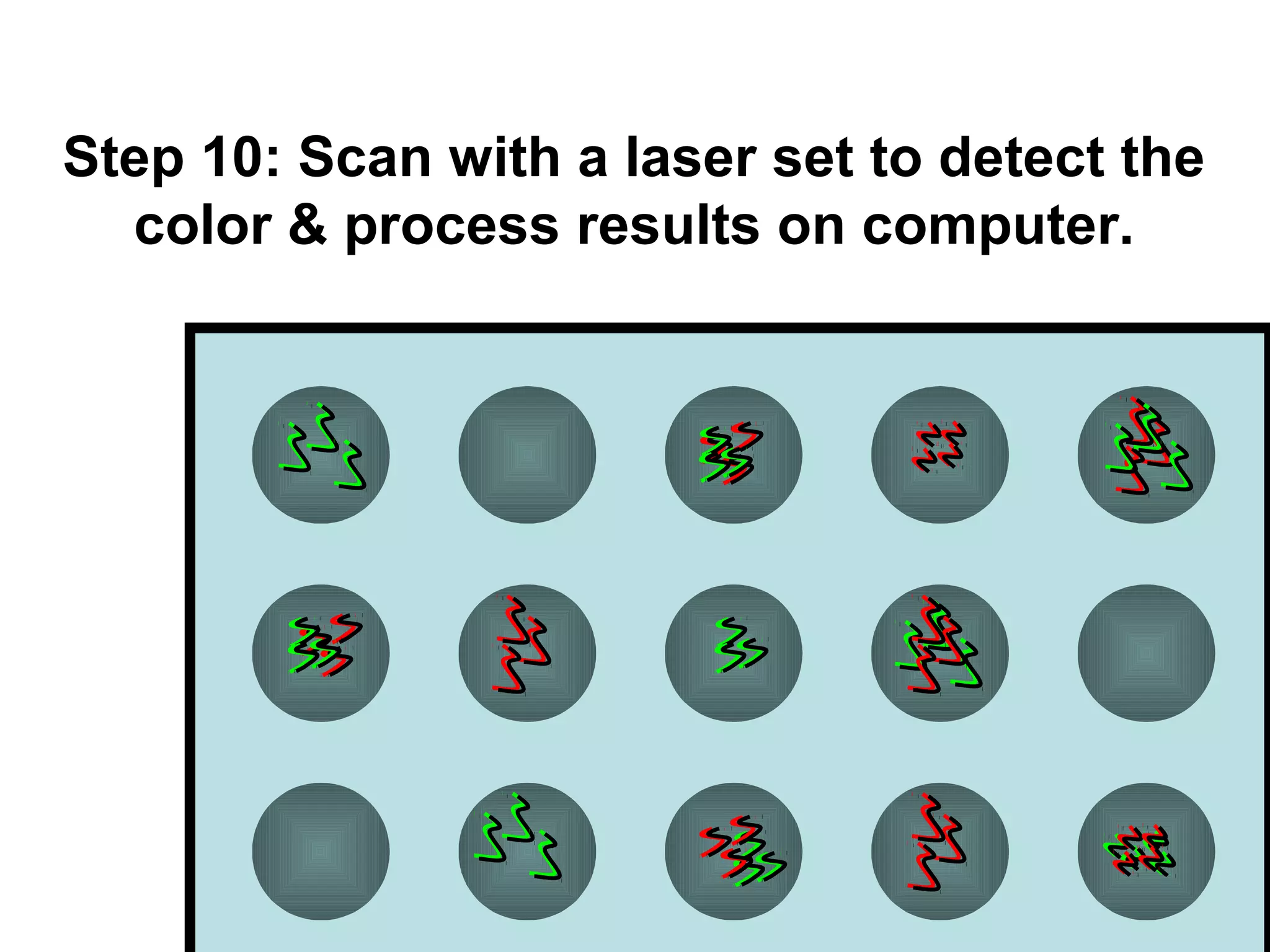
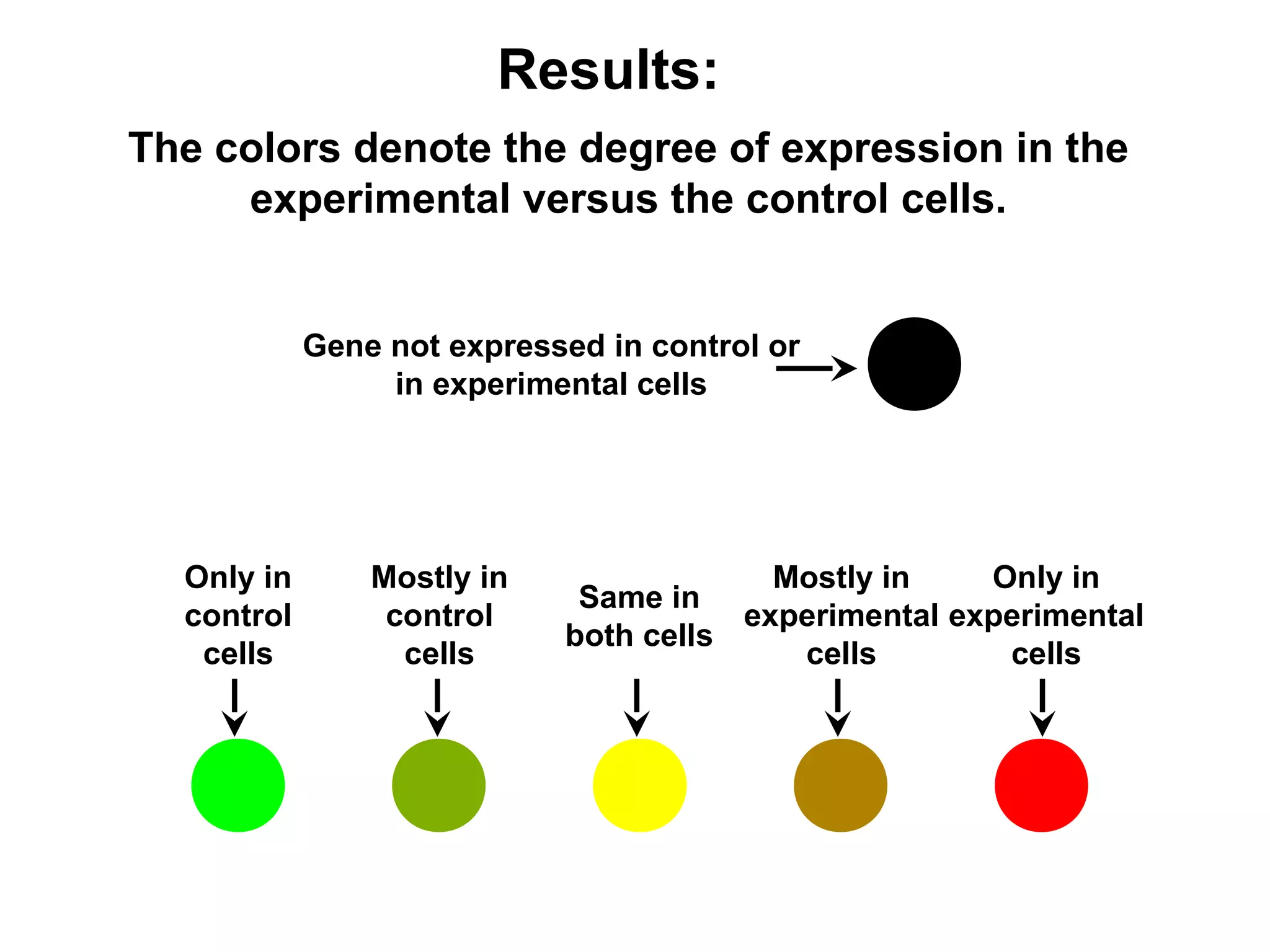
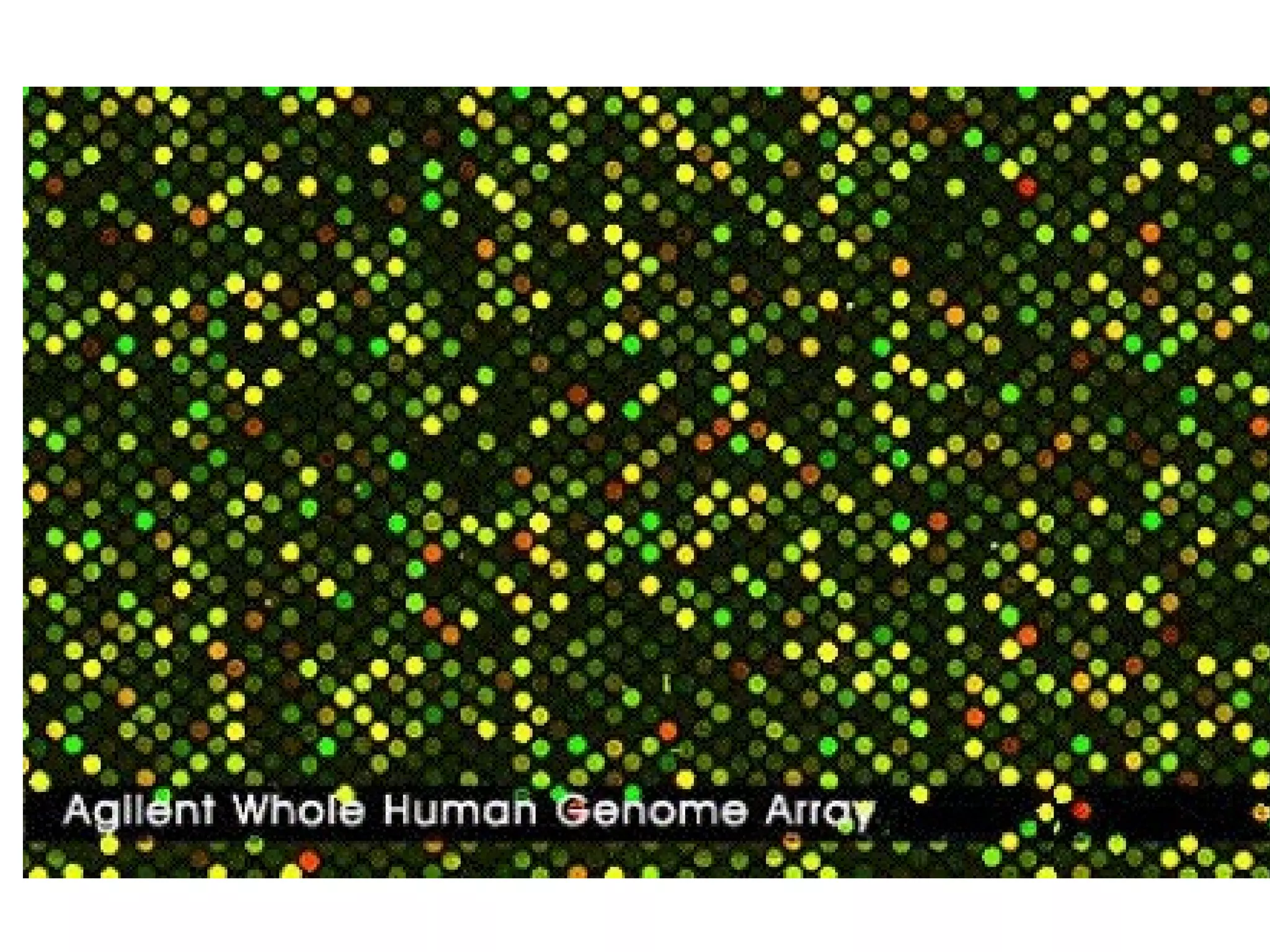
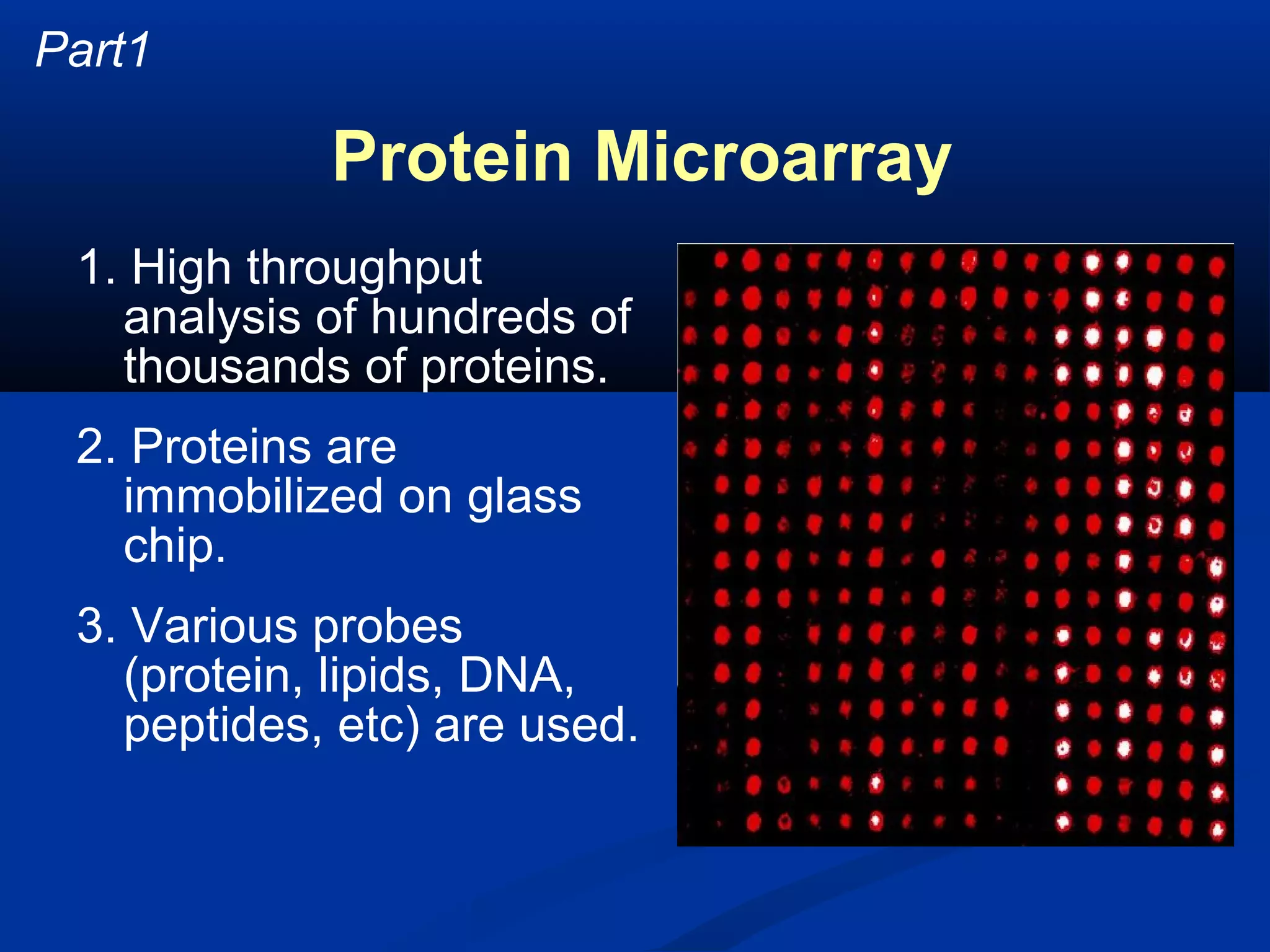
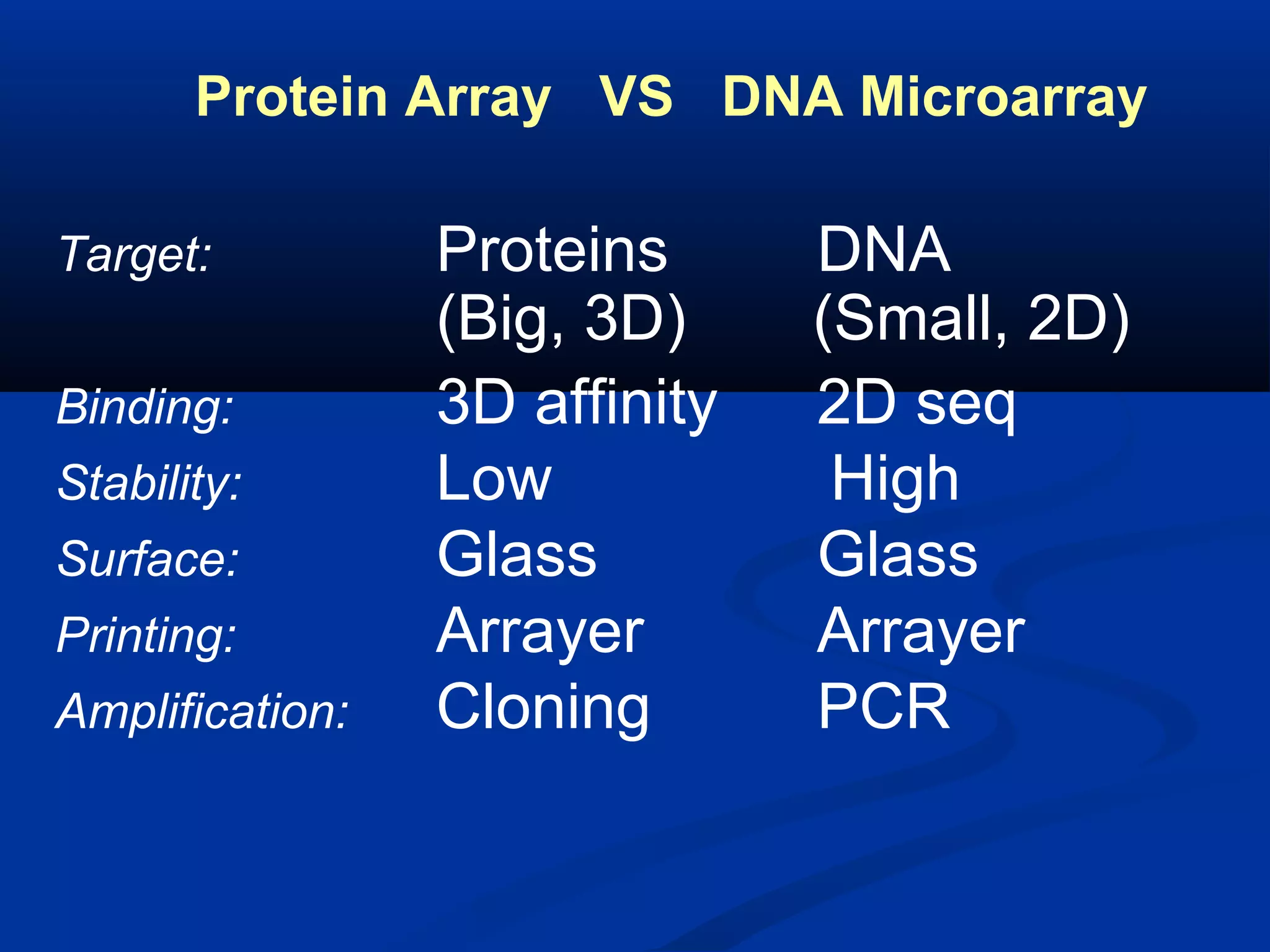
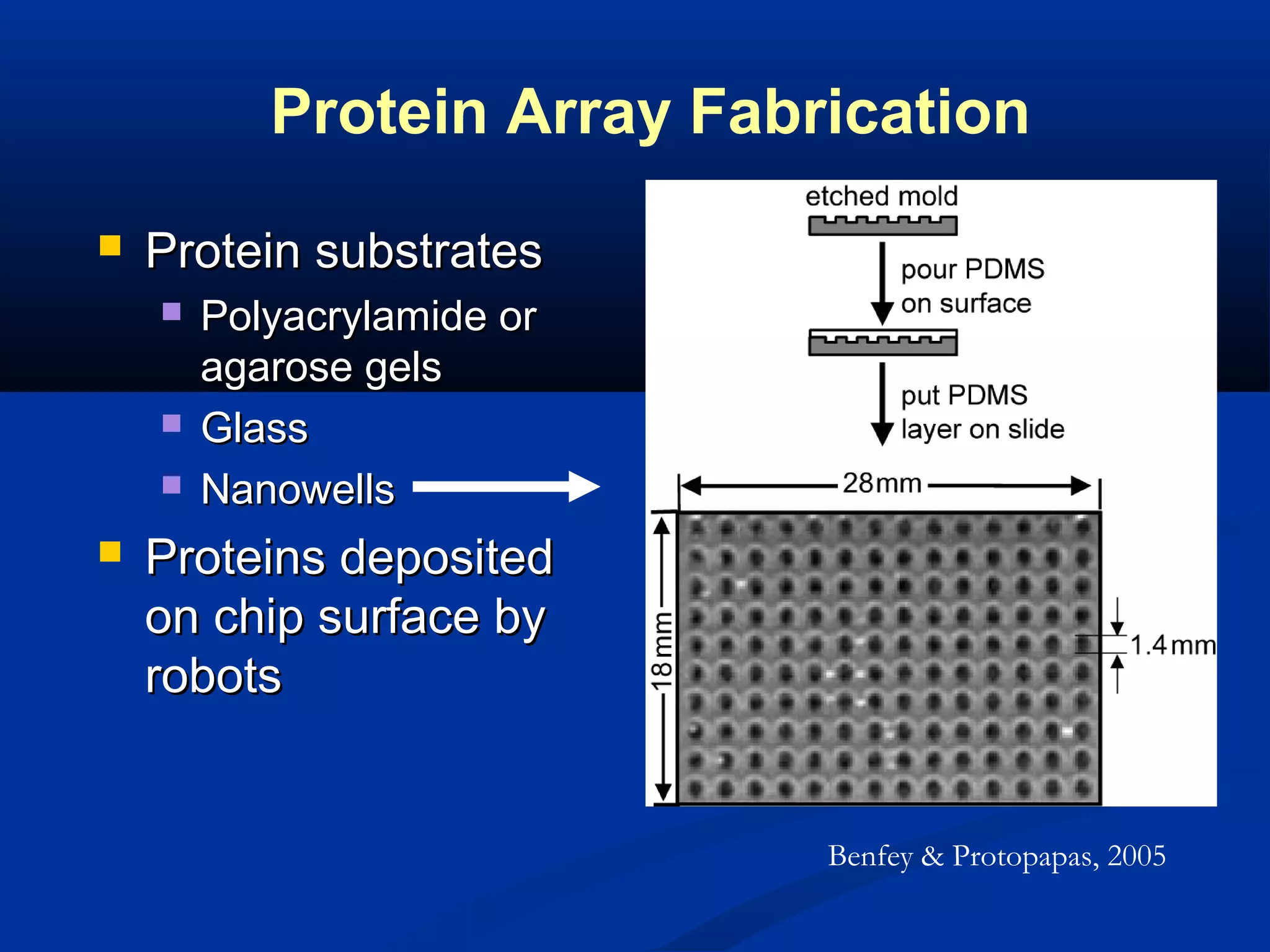
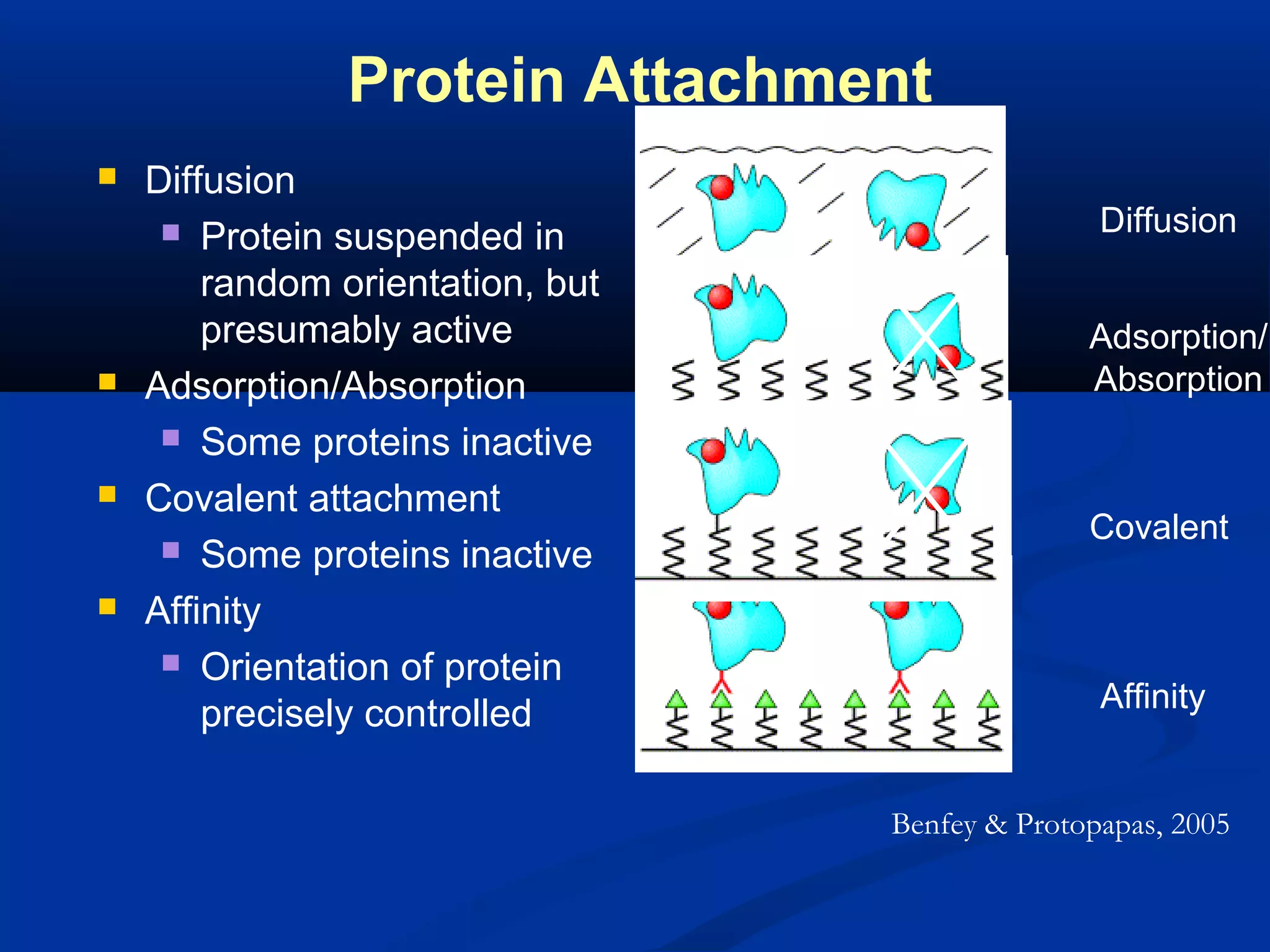
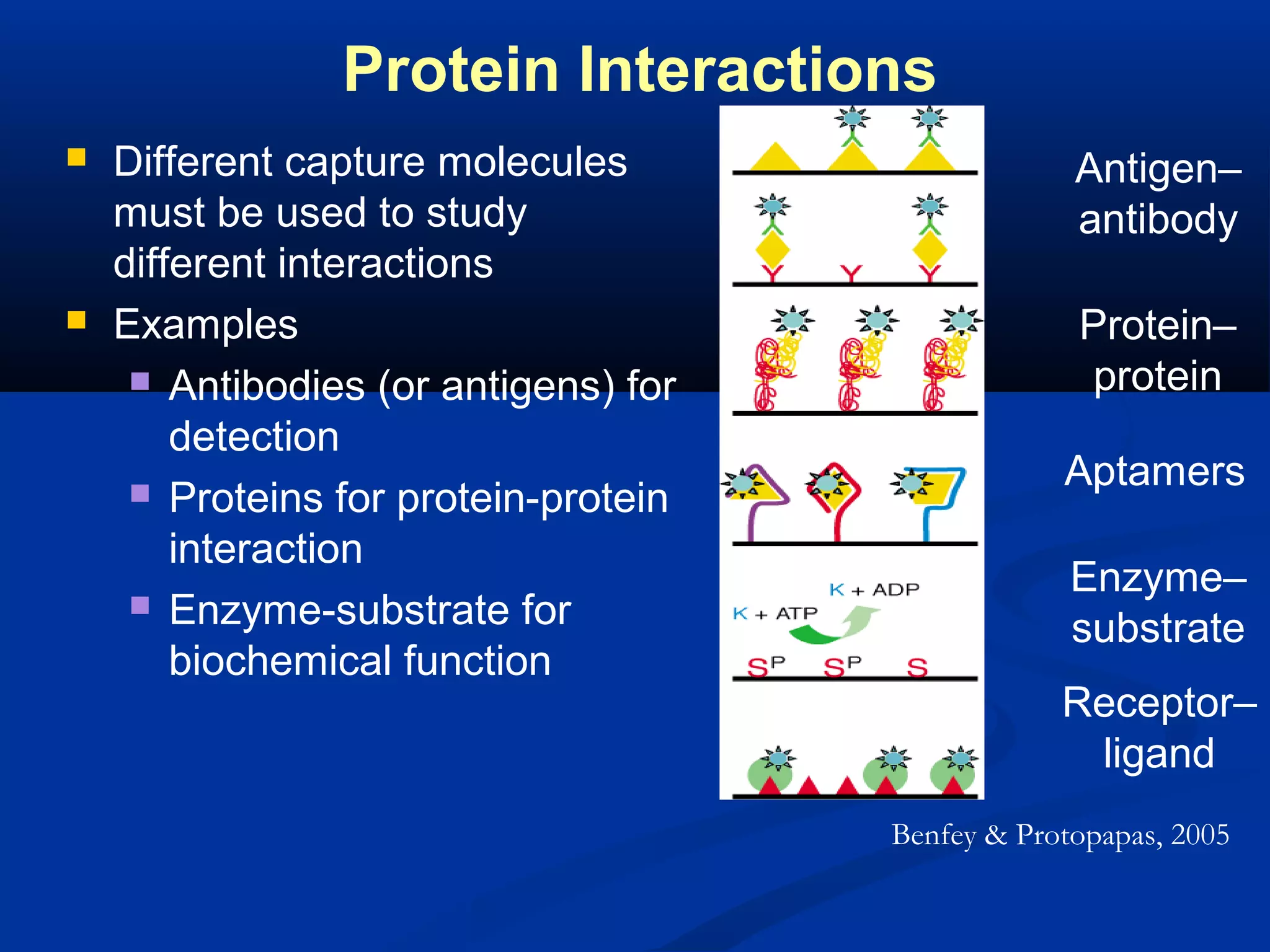
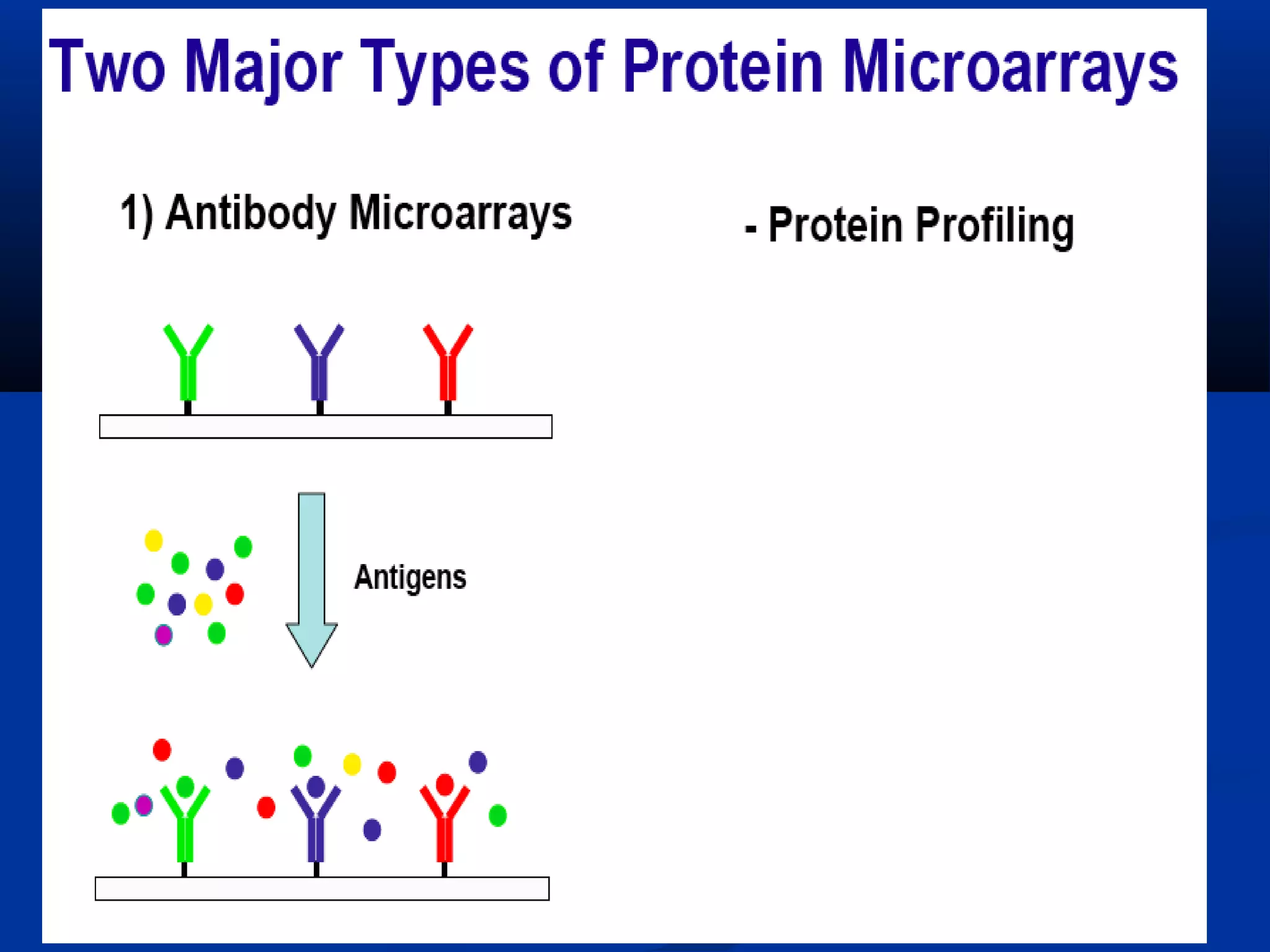
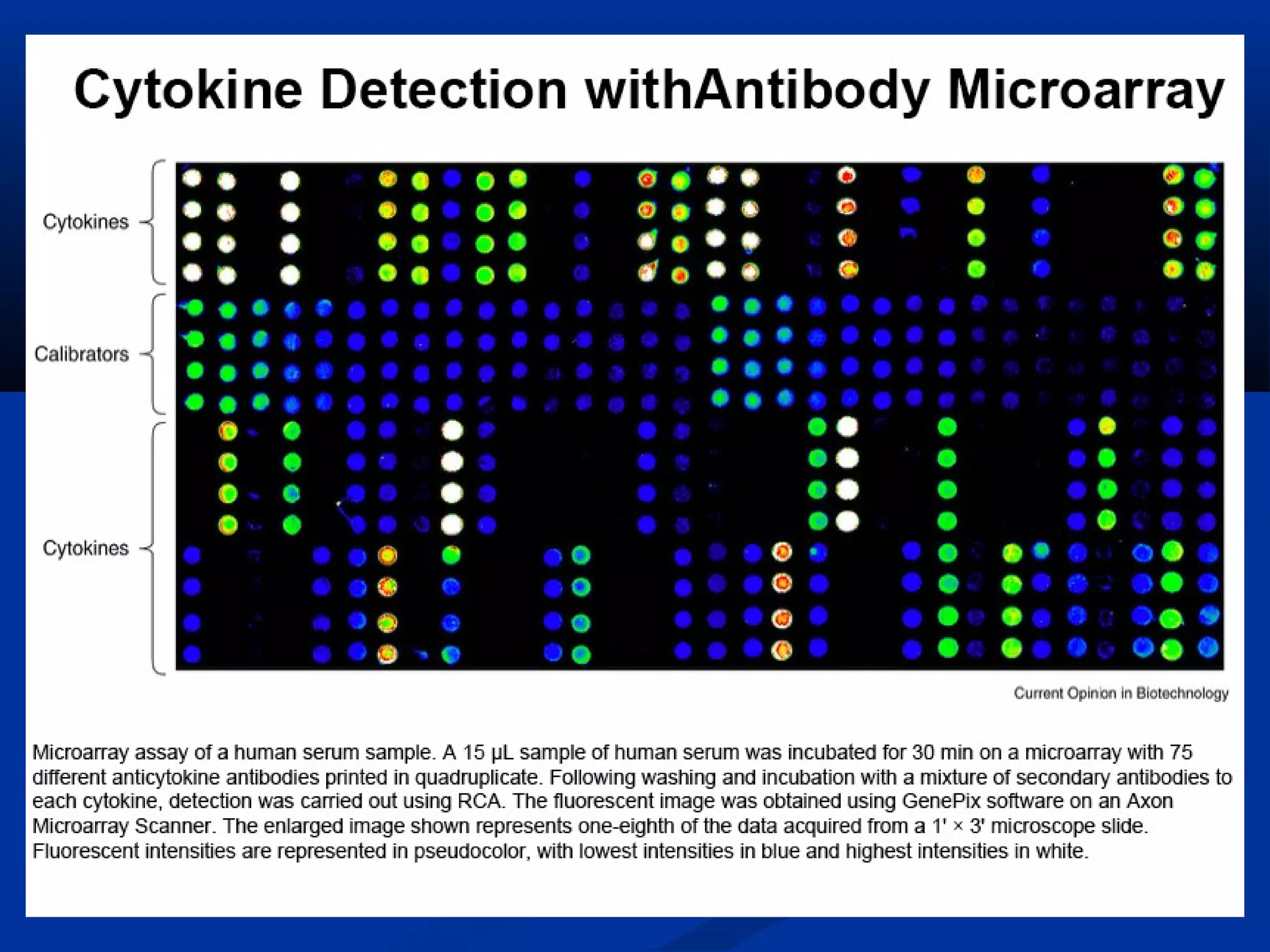
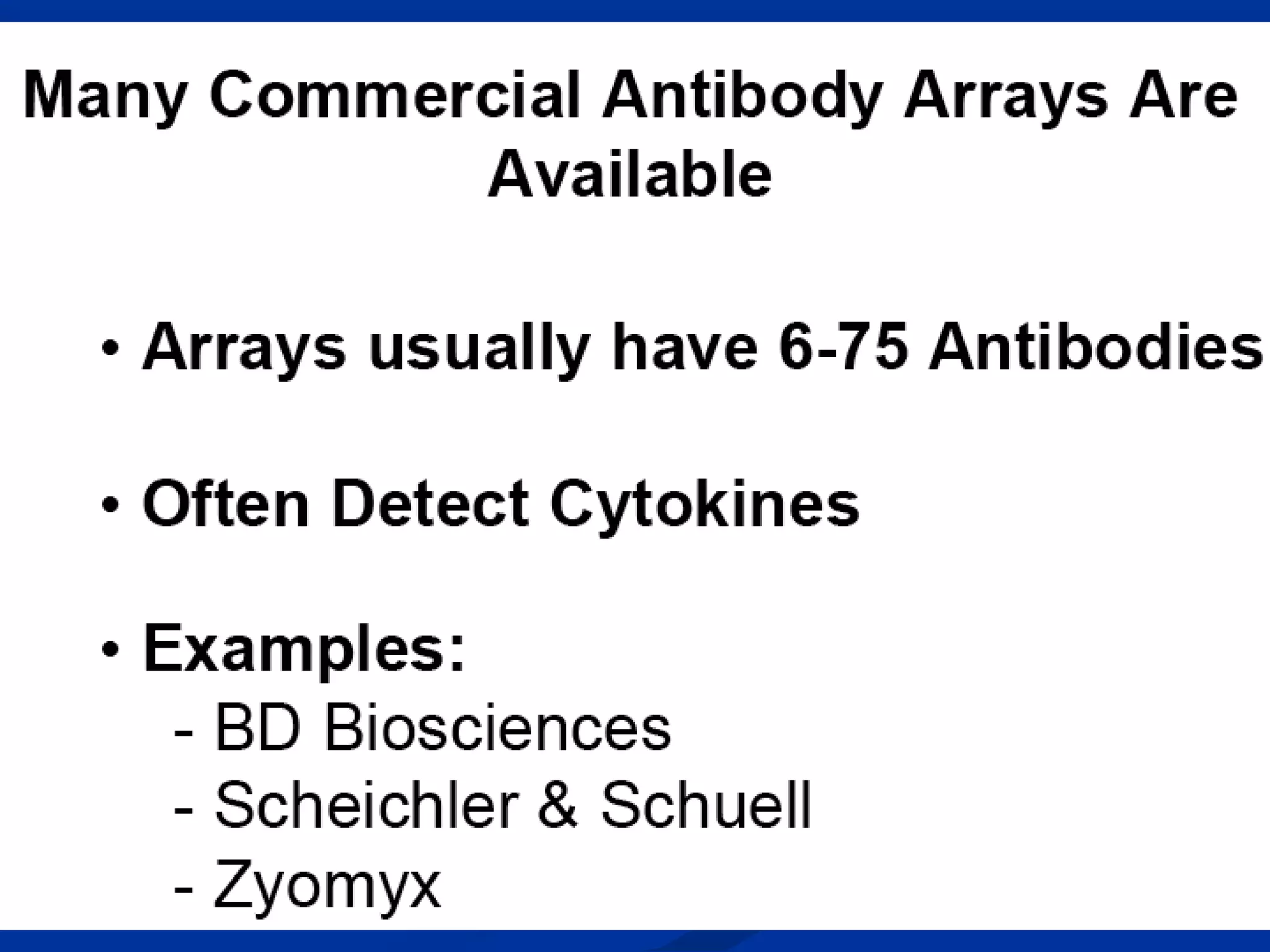
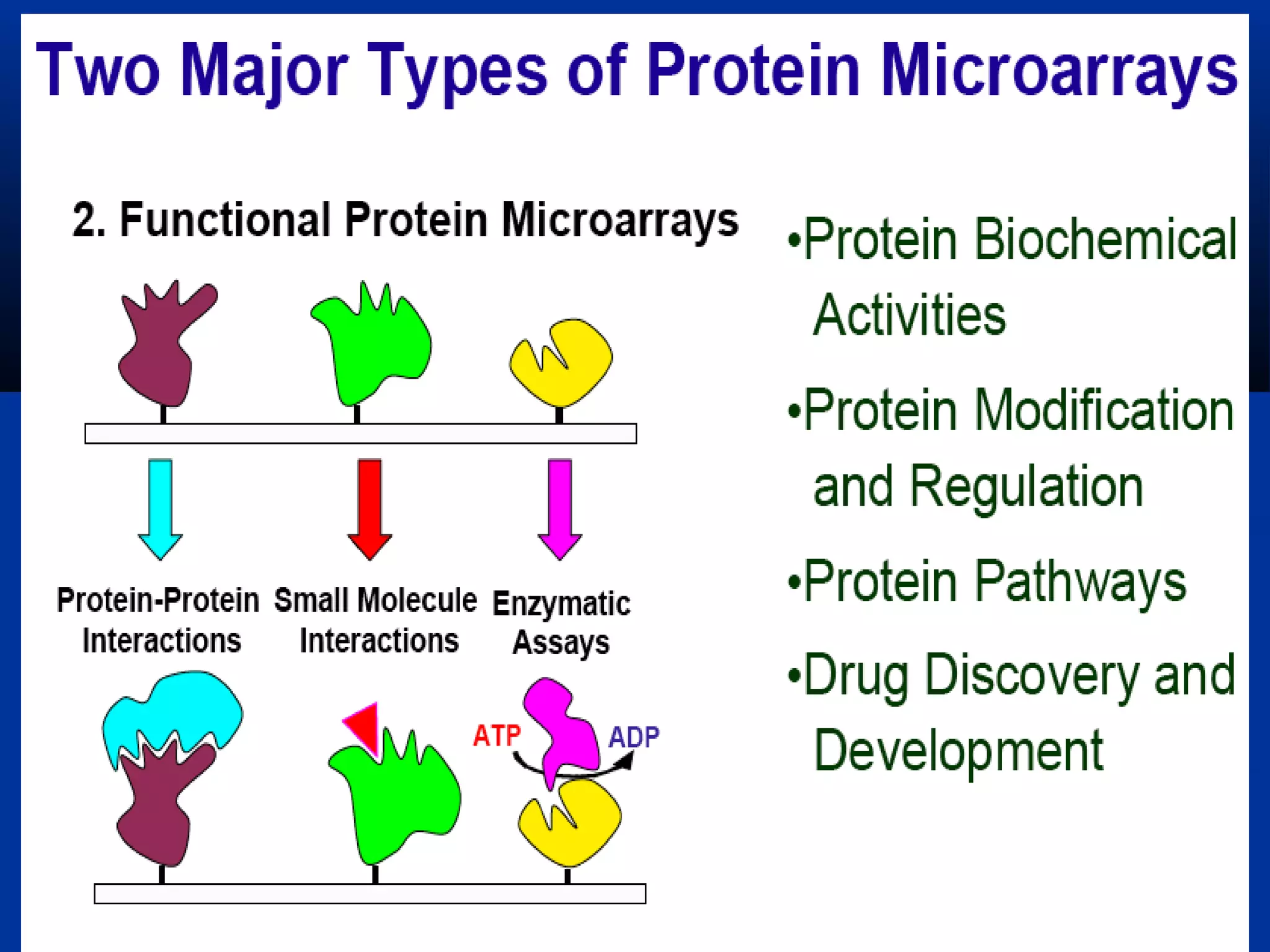
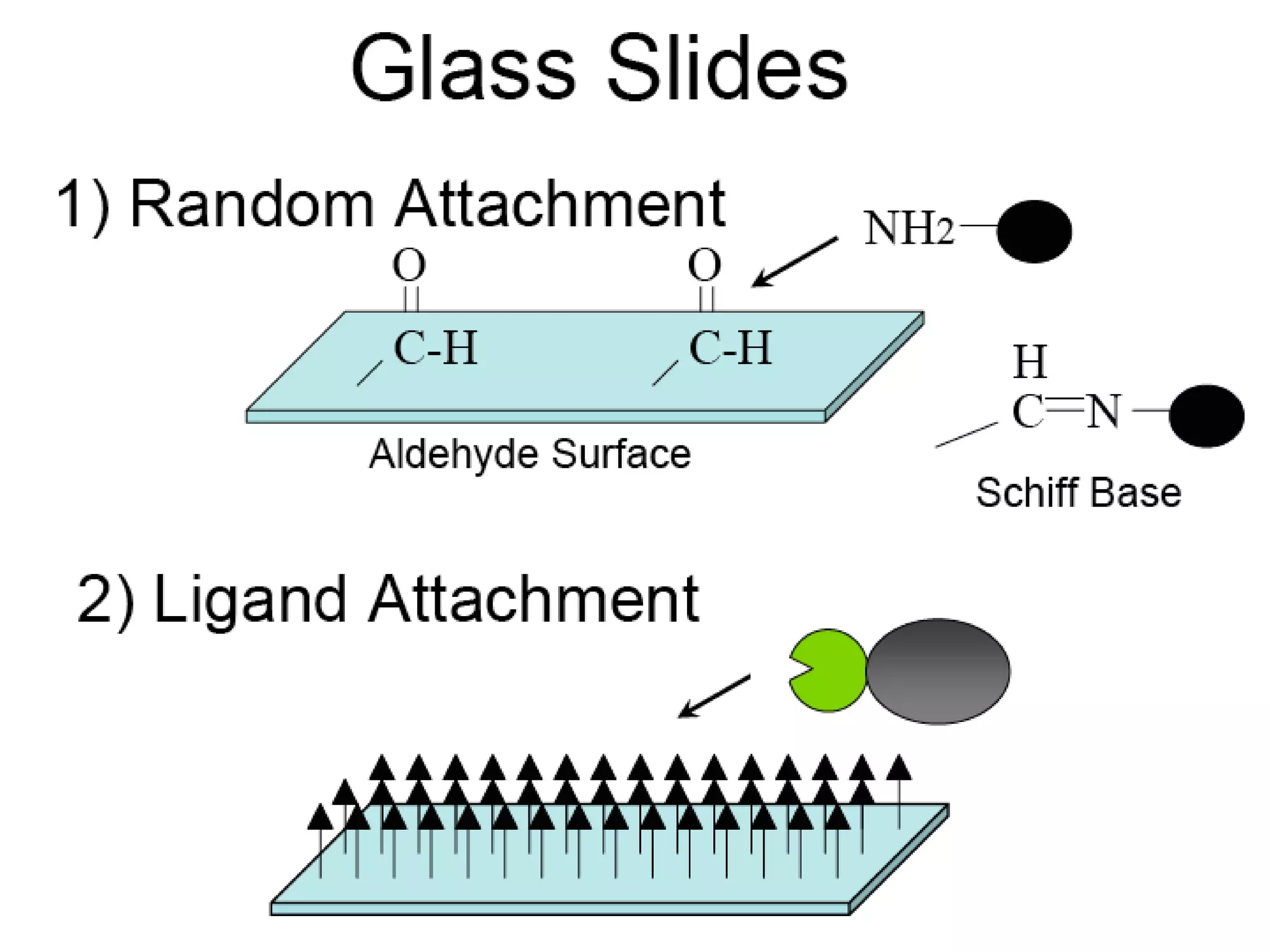
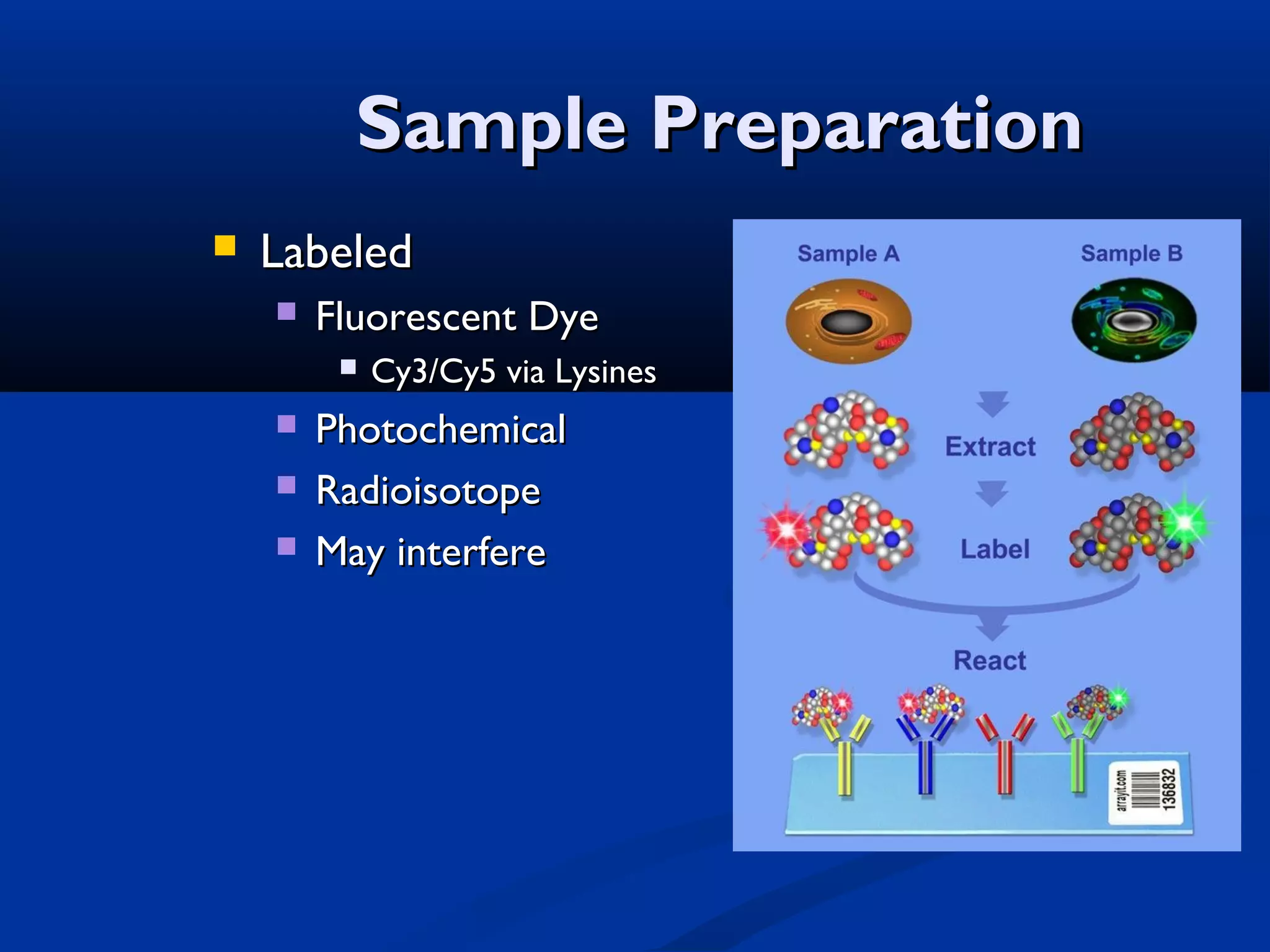
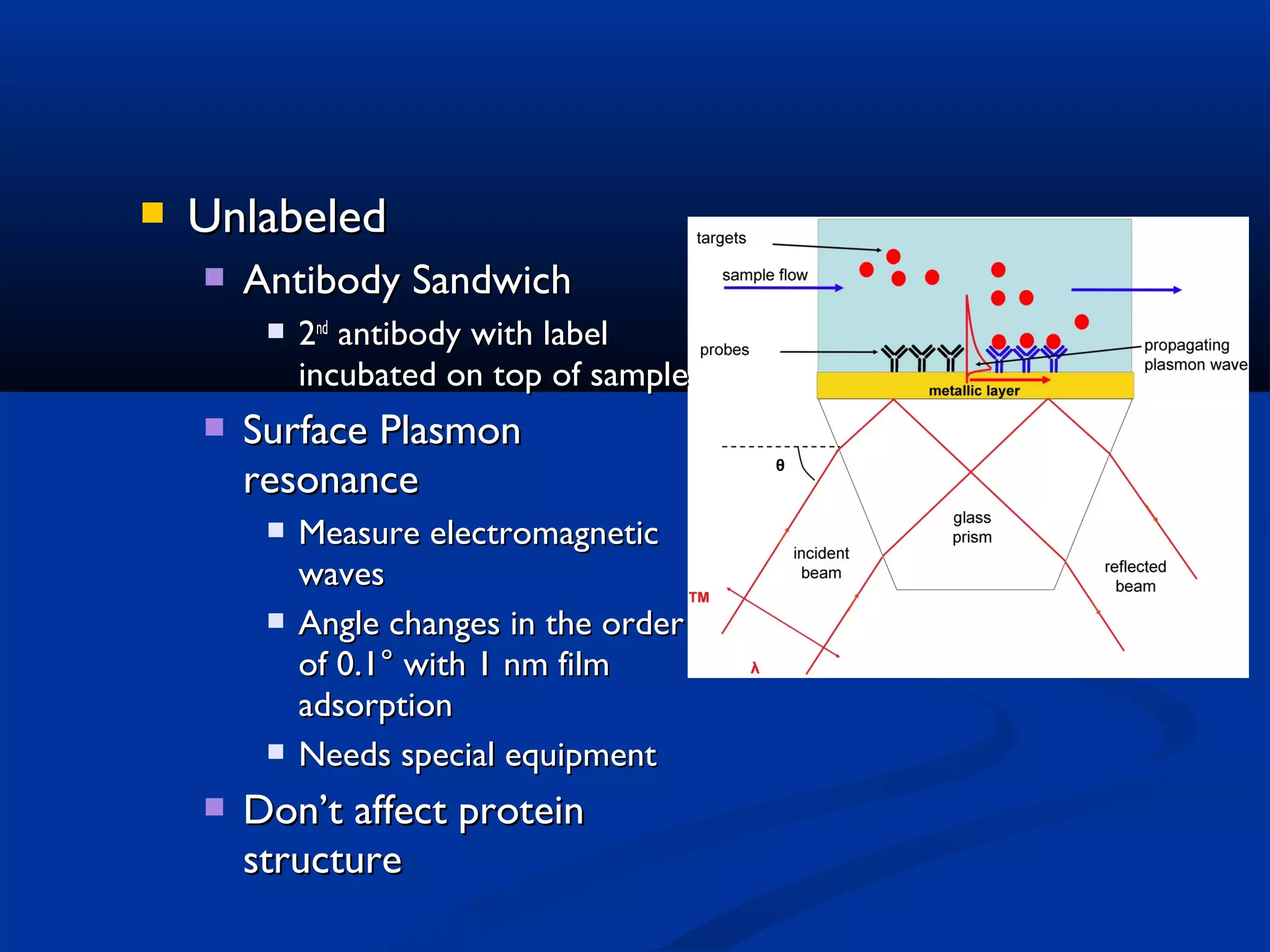
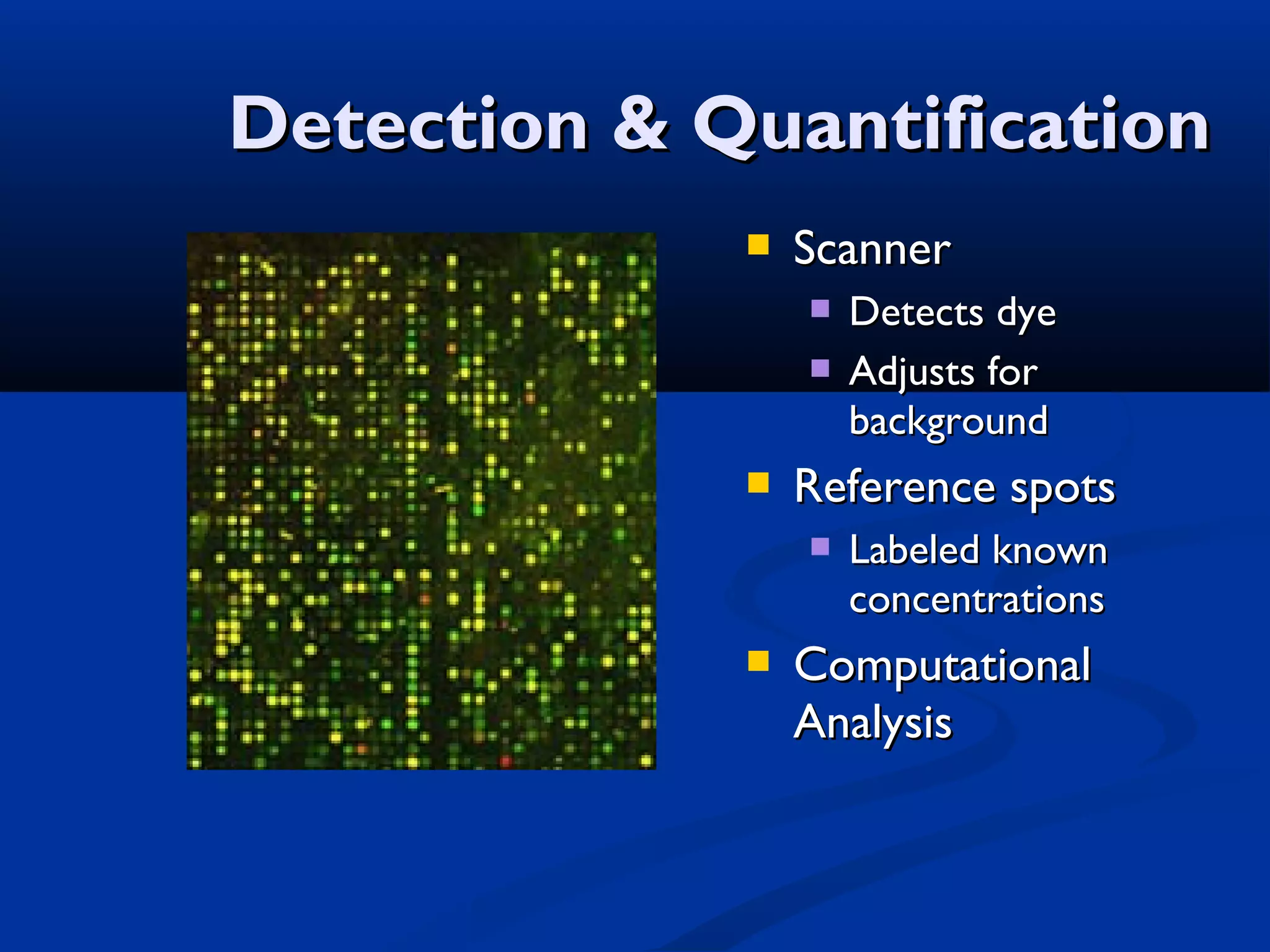
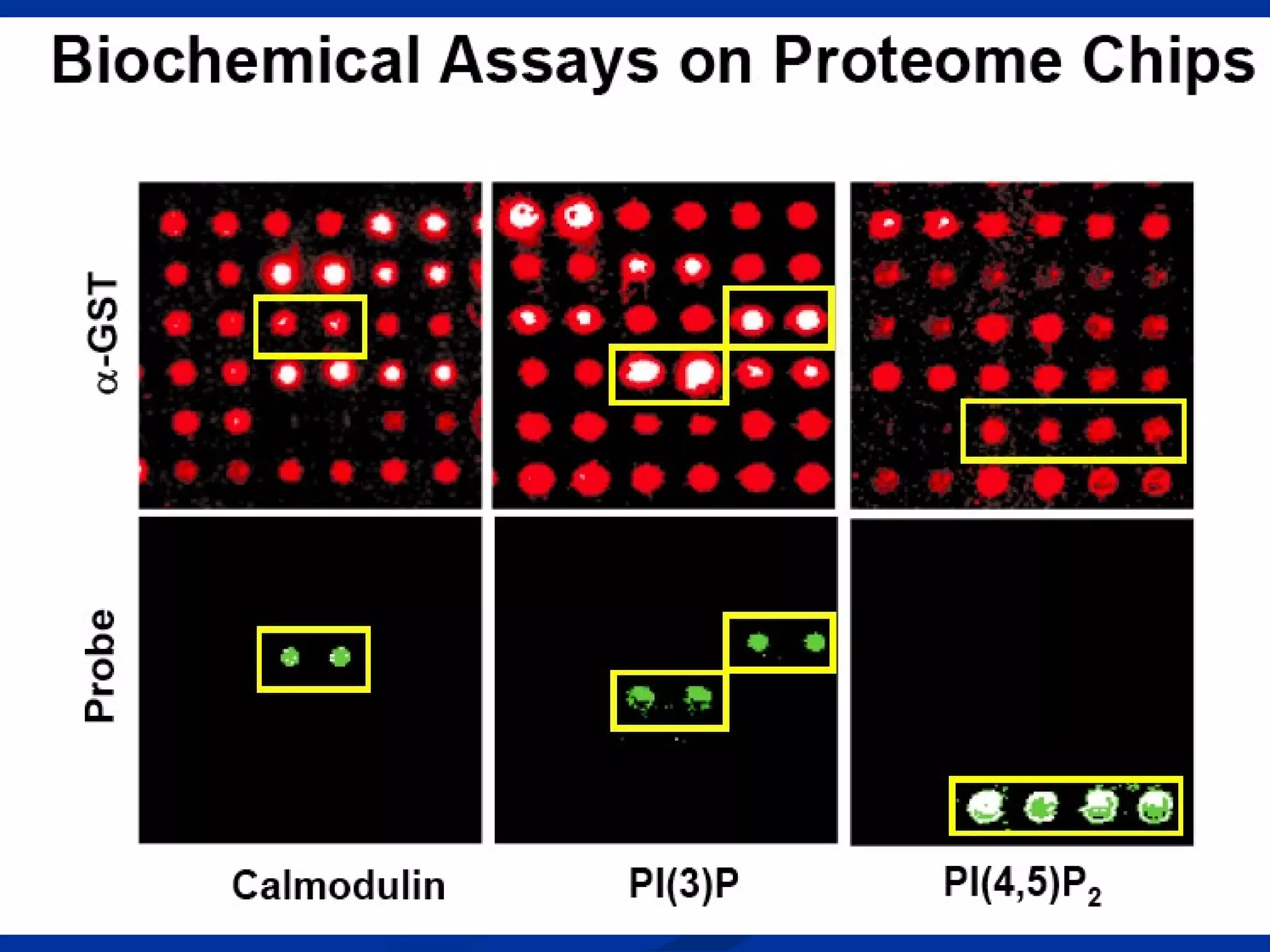
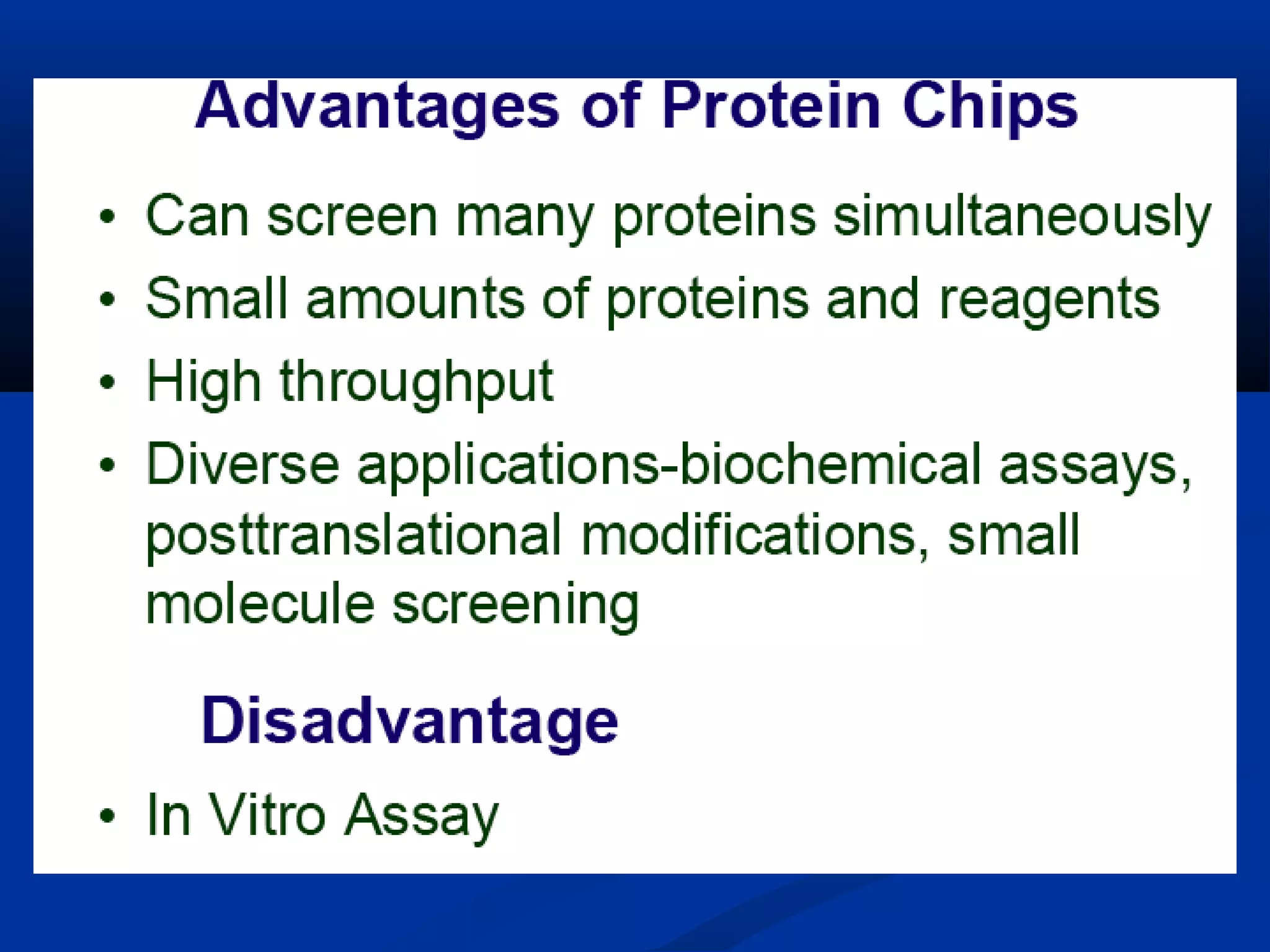
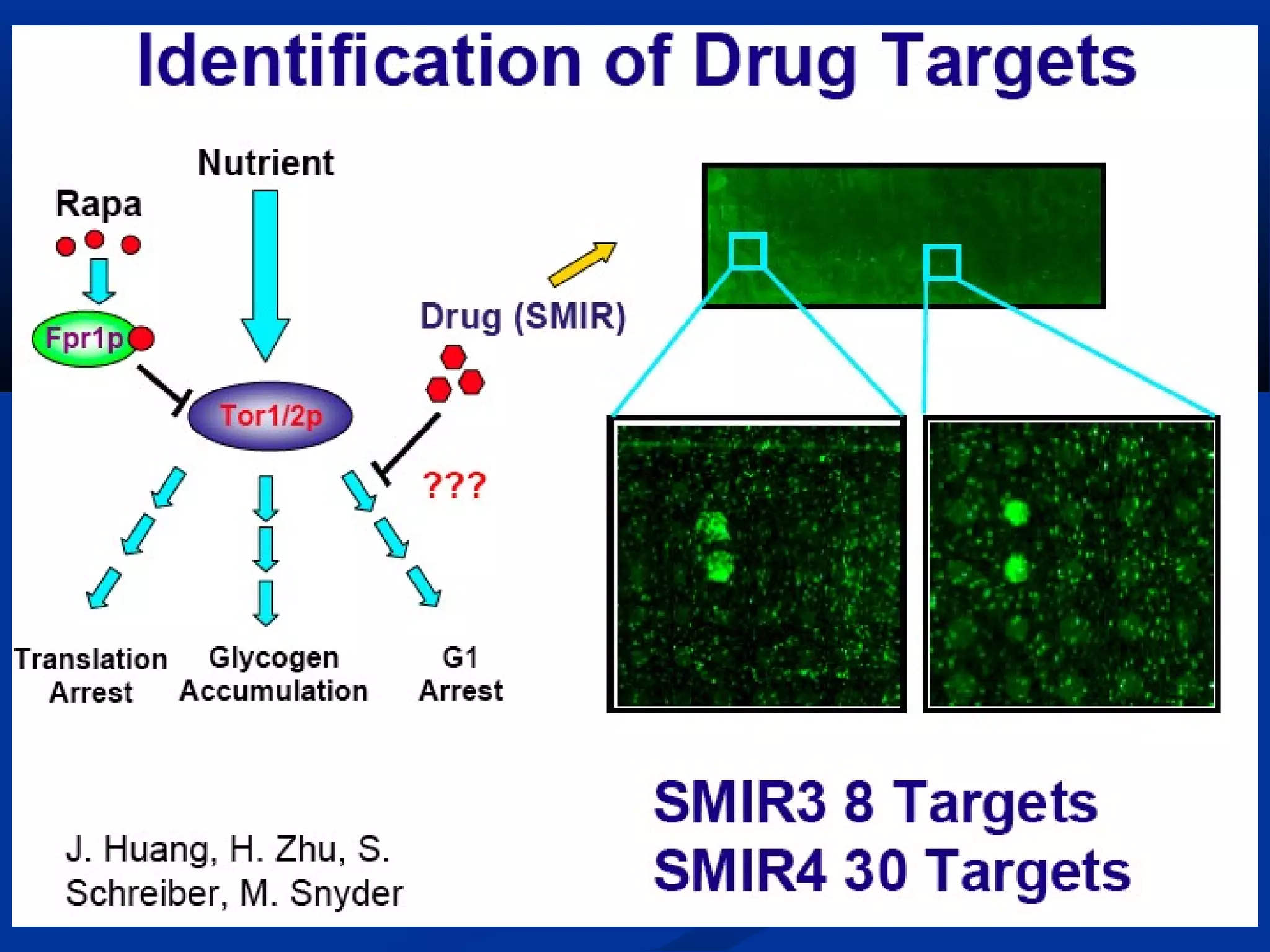
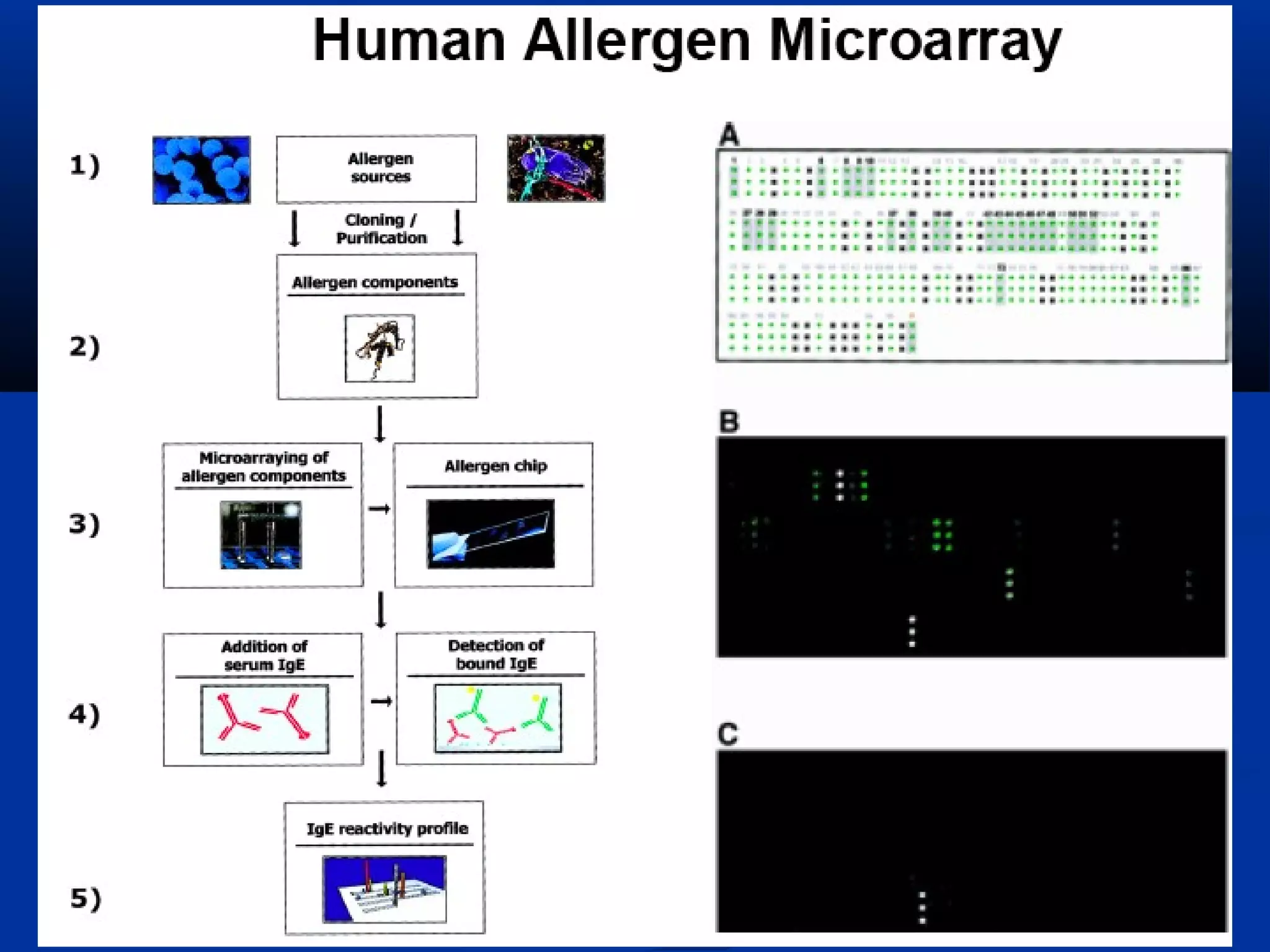
Bio-chips, also known as lab-on-a-chip devices, can provide portable, low-cost, and low-power platforms for integrating sensors and other components. DNA microarrays allow high-throughput screening by placing probes for thousands of genes on a single chip. mRNA is extracted from experimental and control samples, converted to fluorescent cDNA, and hybridized on the chip. The resulting colors indicate gene expression levels. Protein microarrays similarly attach thousands of proteins to a chip and use probes to study protein interactions, expression profiles, and biochemical functions through detection of reaction products. Technical challenges include maintaining protein activity and structure during immobilization and detection.