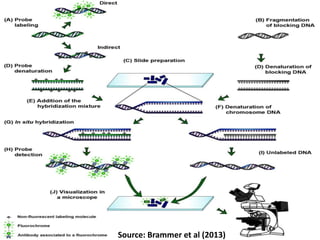
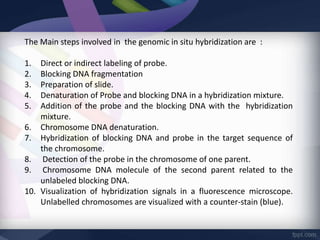
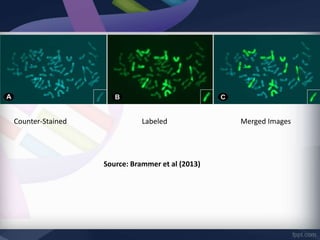
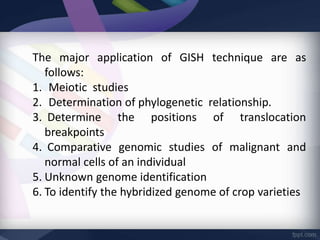
Genomic in situ hybridization (GISH) is a cytogenetic technique that allows radiolabeling of genomic DNA within cells, allowing visualization under a fluorescence microscope. GISH was developed in 1986 for animal hybrid cell lines and 1987 for plant studies. It involves extracting and radiolabeling genomic DNA from one organism as a probe to target and hybridize to similar genomic regions of another organism. Unhybridized regions can then be stained, allowing visualization of probe-target complexes and unlabeled regions. GISH provides quick, sensitive, and informative results for establishing phylogenetic relationships and identifying hybridized genomes.