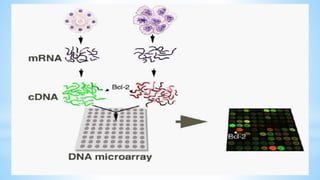
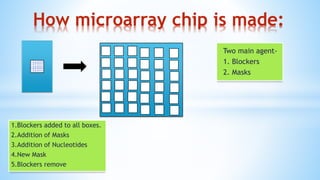
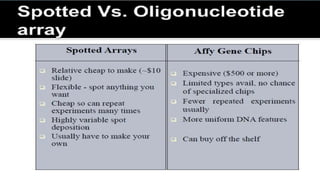
Biplob Dey presents on DNA microarray technique. A DNA microarray is a collection of DNA spots attached to a solid surface, with each known gene occupying a particular spot. Fluorescently labeled target sequences that bind to probe sequences generate a signal, showing levels of gene activity. DNA microarrays allow samples to be labeled with fluorescent dyes and hybridized to a chip to detect complementary nucleic acid sequences through hydrogen bonding and measure gene expression patterns. There are different types of microarrays including spotted DNA arrays and oligonucleotide arrays. Applications include detecting cancer cells and studying antibiotic activity.