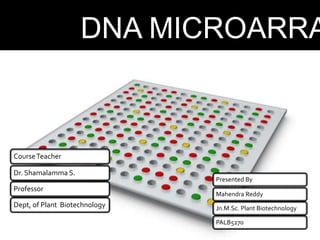
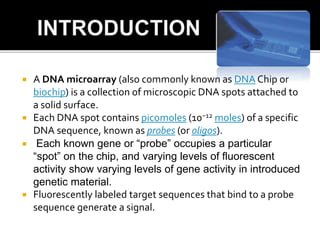
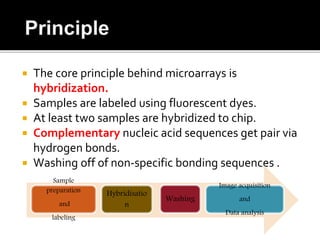
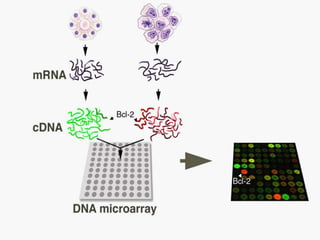
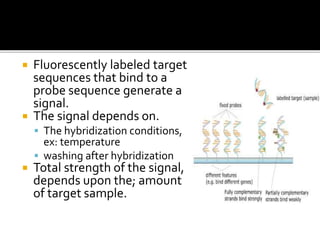
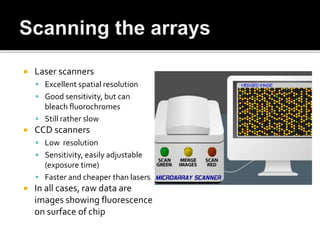
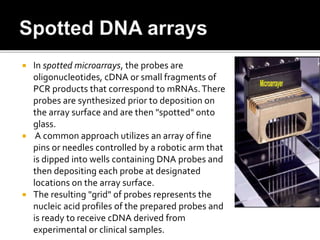
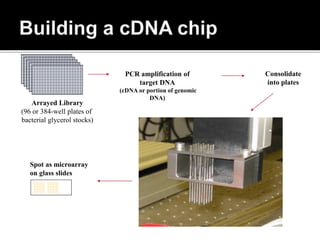
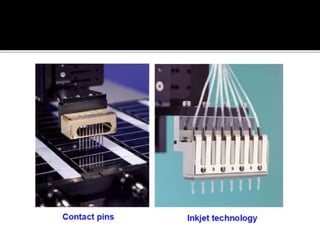
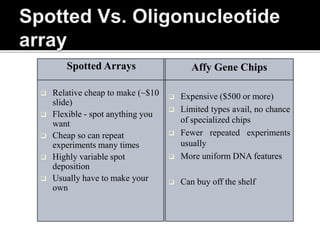
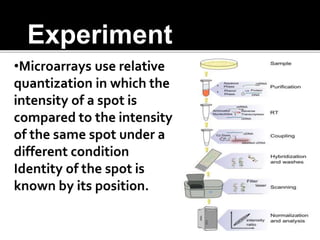
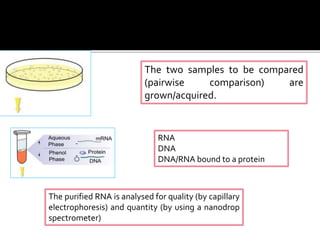
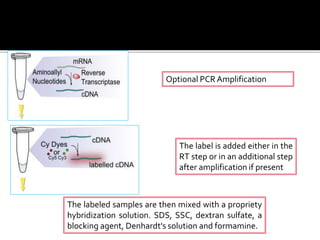
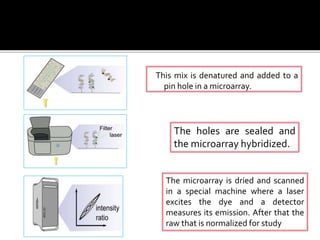
Dr. Shamalamma S. presented on DNA microarrays. DNA microarrays allow thousands of genes to be compared simultaneously by attaching DNA probes to a chip which fluorescently labeled samples can bind to. The chip is then scanned to analyze gene expression levels. Applications include disease diagnosis, toxicology studies, and pharmacogenomics. While a powerful tool, microarrays have limitations such as lack of knowledge about many genes and lack of standardization.