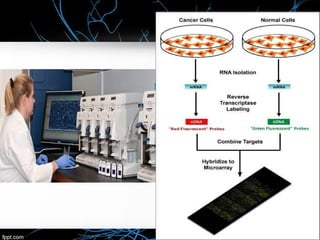
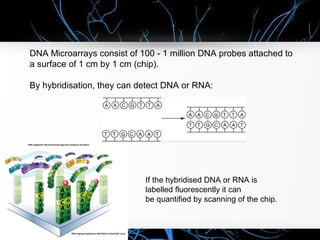
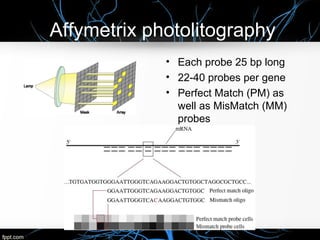
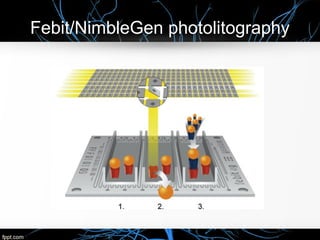
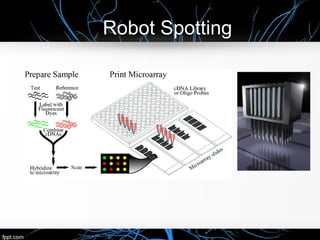
This document discusses DNA microarray technology, which allows researchers to study thousands of genes simultaneously using a chip that contains DNA probes. It describes how DNA microarray works by hybridizing labeled DNA or RNA samples to the probes on the chip to detect gene expression patterns. Different types of microarrays like expression analysis, mutation analysis, and comparative genomic hybridization are used for various applications such as gene discovery, disease diagnosis, drug discovery, and toxicological research.