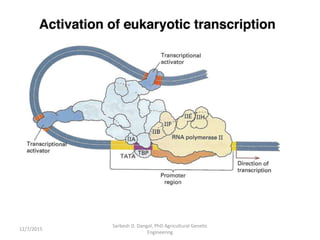
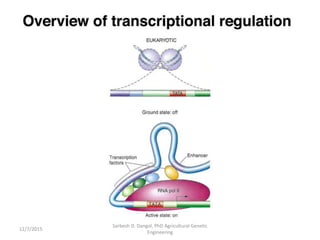
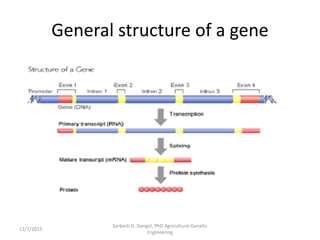
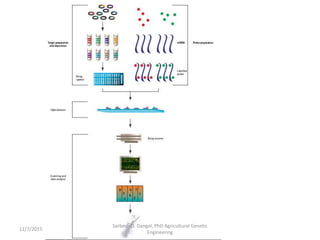
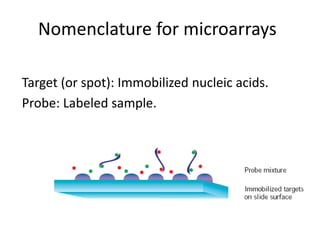
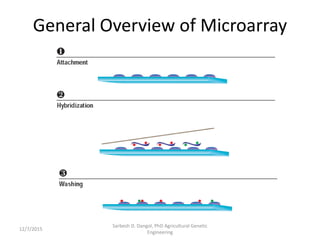
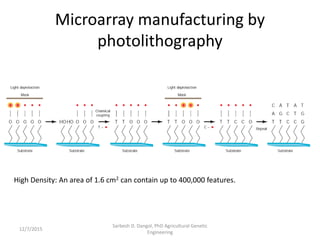
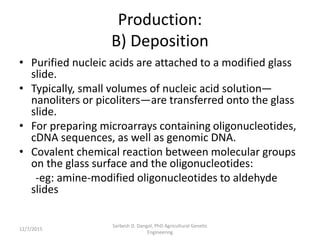
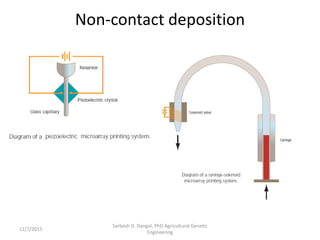
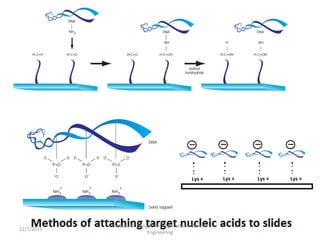
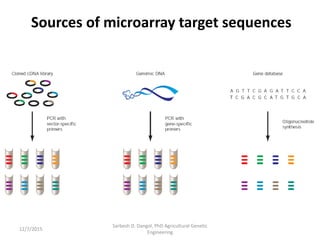
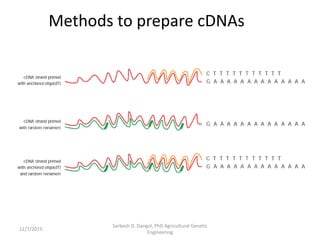
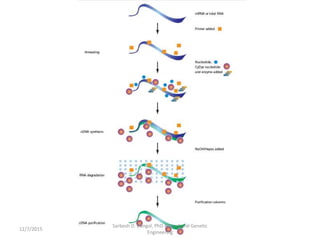
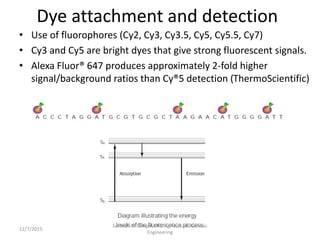
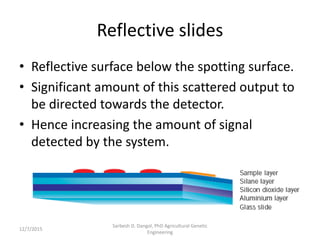
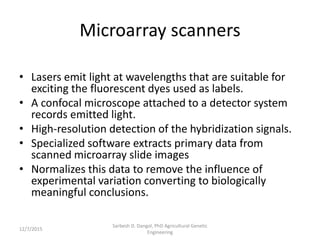
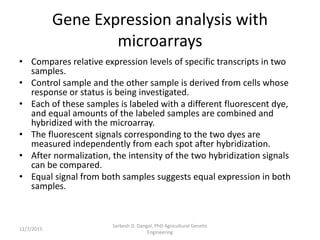
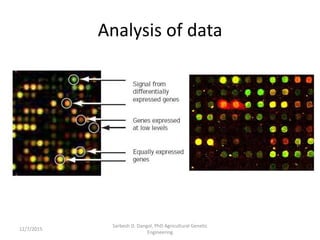
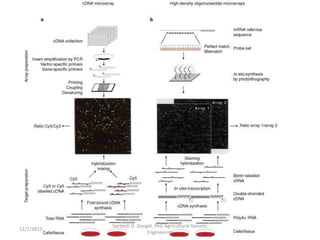
The document provides an introduction to microarrays. It describes that microarrays allow for the simultaneous assessment of large numbers of nucleic acids in parallel using molecular hybridization methods. Microarrays involve preparing miniaturized collections of known nucleic acid sequences that are immobilized on a solid surface as targets. Labeled mRNA or DNA samples are used as probes to determine expression levels of thousands of genes at once through detecting which probes hybridize to which targets. The document outlines the basic components and manufacturing of microarrays as well as their applications in gene expression analysis, genome analysis, and drug discovery.