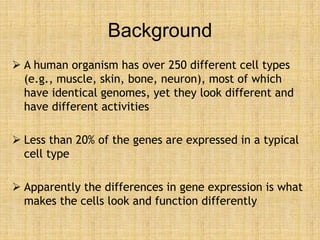
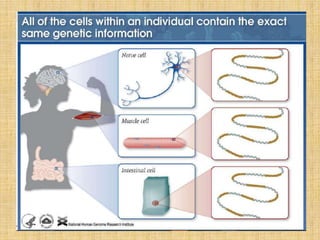
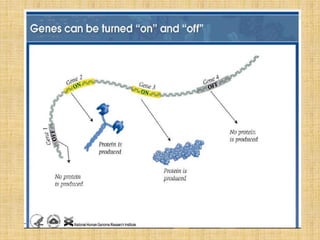
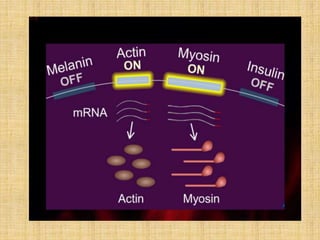
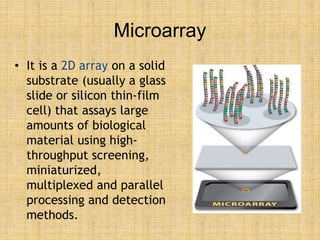
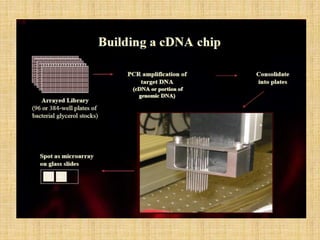
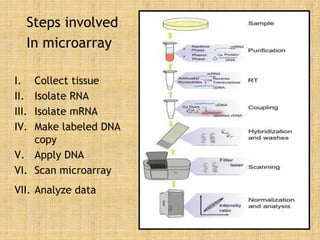
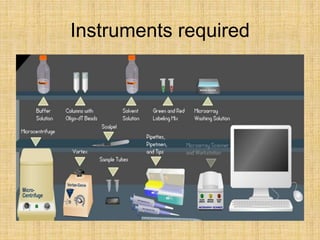
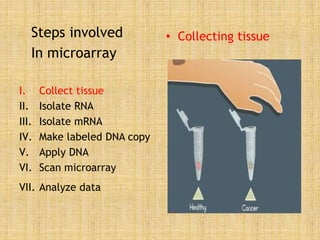
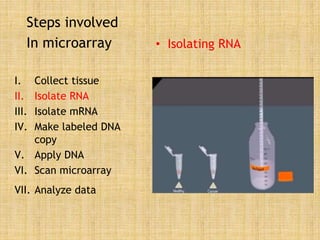
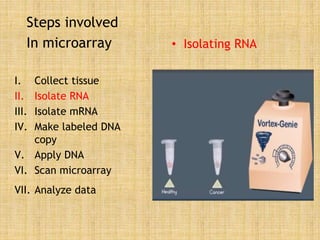
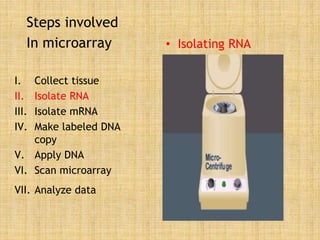
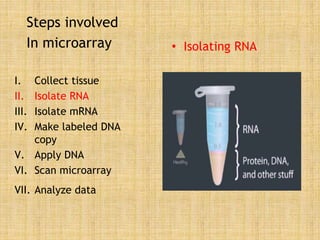
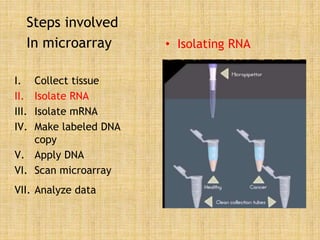
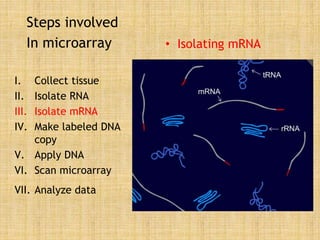
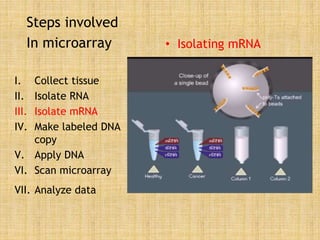
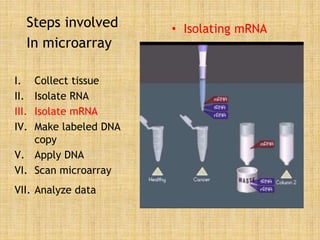
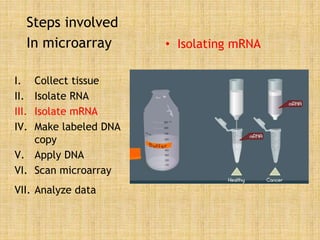
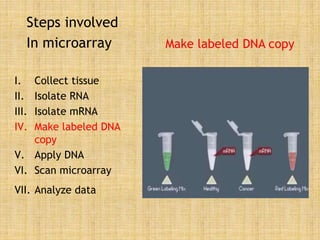
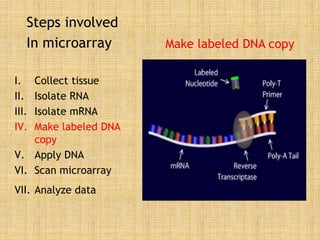
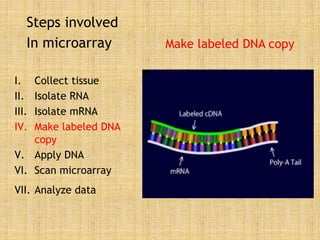
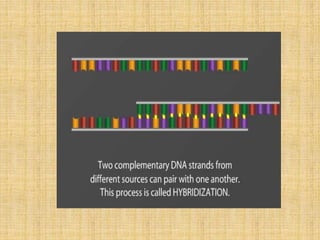
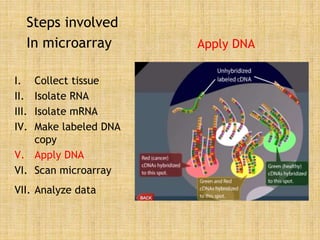
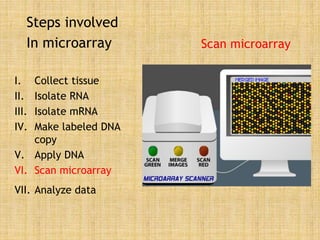
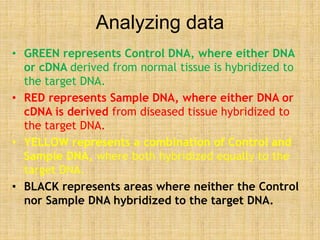
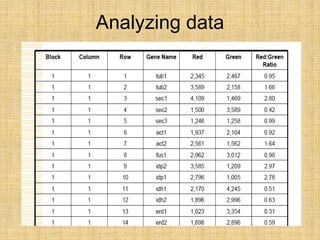
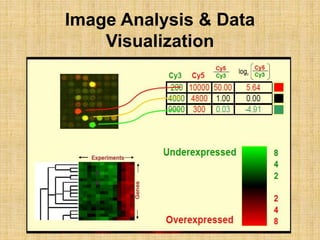
Microarrays allow researchers to study gene expression across thousands of genes simultaneously. They work by hybridizing labeled cDNA or cRNA to probes attached to a solid surface, then detecting and quantifying the hybridized genes. The document outlines the history and development of microarray technology. It describes the key steps in a DNA microarray experiment including tissue collection, RNA isolation, cDNA synthesis, hybridization to the array, scanning, and data analysis. Applications include studying gene expression in health and disease, drug development, and pharmacogenomics. Advantages are the ability to study many genes at once, while limitations include expense and complexity of data analysis.