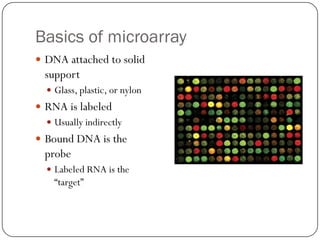
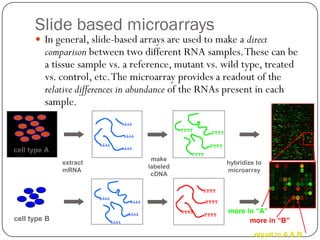
This document outlines the principles and applications of DNA microarrays, which are used to measure gene expression and genetic variations by hybridizing DNA sequences on a solid surface. It discusses different types of microarrays, including cDNA and oligonucleotide microarrays, and their historical development from Southern and Northern blotting techniques. The document also highlights various applications such as gene expression profiling, SNP analysis, and comparative genomic hybridization.