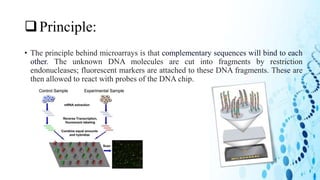
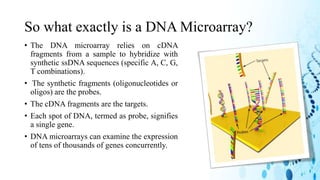
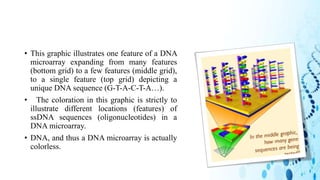
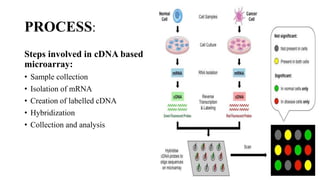
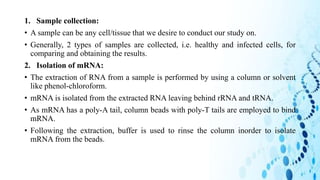
This document discusses DNA microarray technology. It begins with an introduction, explaining that DNA microarrays allow analysis of thousands of genes simultaneously through hybridization of fluorescently labeled DNA probes to a microarray slide. It then covers the principles of DNA microarrays, including the types (cDNA and oligonucleotide), how they are constructed with probes immobilized on a solid surface, and how hybridization allows analysis of gene expression profiles. Applications discussed include drug discovery, disease diagnostics, and functional genomics. Advantages are high-throughput analysis and ability to study many genes, while disadvantages include potential for false results from single experiments.