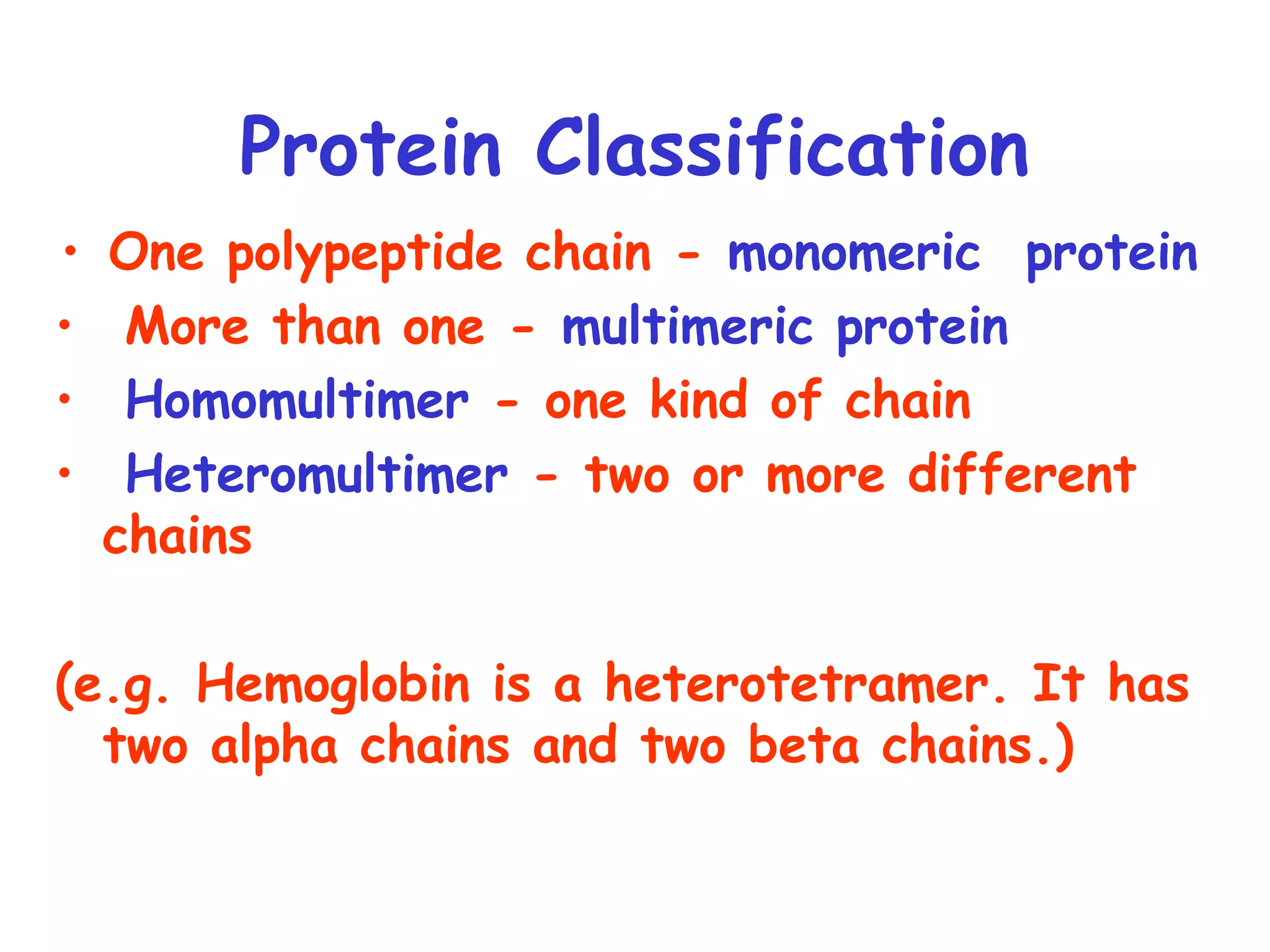
Chapter 4 covers protein structure and function, detailing terminology such as conformation and protein classification into simple, conjugated, monomeric, and multimeric categories. It discusses the four levels of protein structure and the significance of non-covalent forces in determining that structure, as well as common secondary structures like alpha helices and beta-sheets. The chapter highlights various protein functions, including catalysis, transport, and structural roles, while addressing the impact of amino acid changes on protein functionality, exemplified by conditions like sickle cell anemia and osteoarthritis.