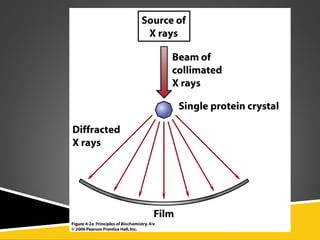
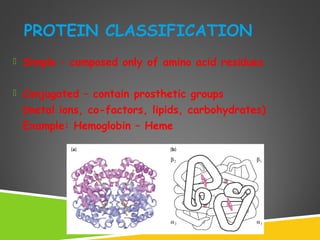
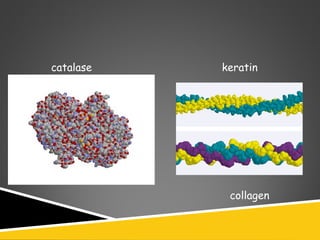
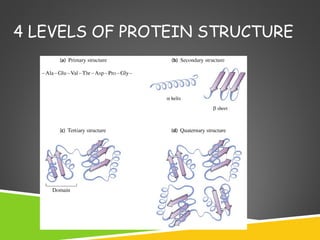
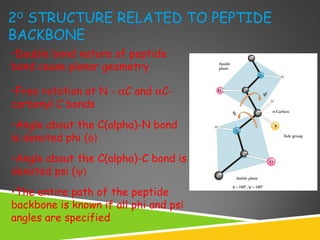
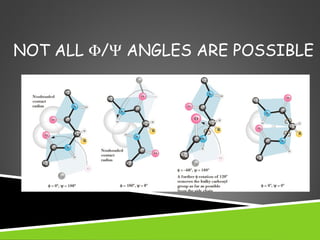
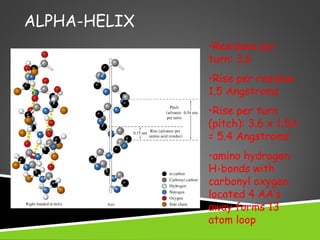
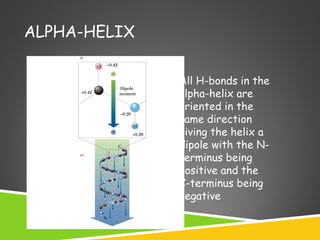
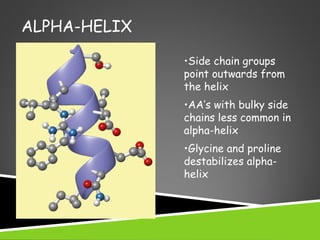
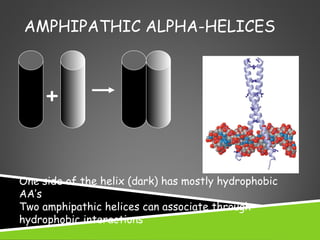
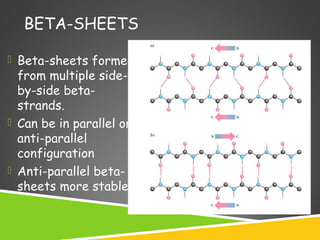
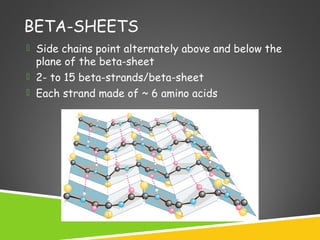
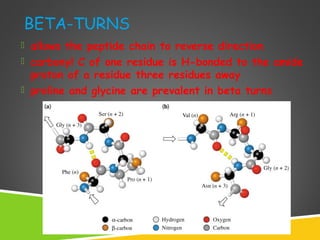
This document discusses protein structure and function. It defines different levels of protein structure from primary to quaternary, and describes common secondary structures like alpha helices and beta sheets. Alpha helices are right-handed coils stabilized by hydrogen bonds between amino acids four positions apart in the sequence. Beta sheets consist of beta strands connected laterally by hydrogen bonds to form flat sheets, which can be arranged in parallel or anti-parallel configurations. Tertiary structure refers to the overall three-dimensional shape of a protein formed by the folding and interaction of secondary structures.