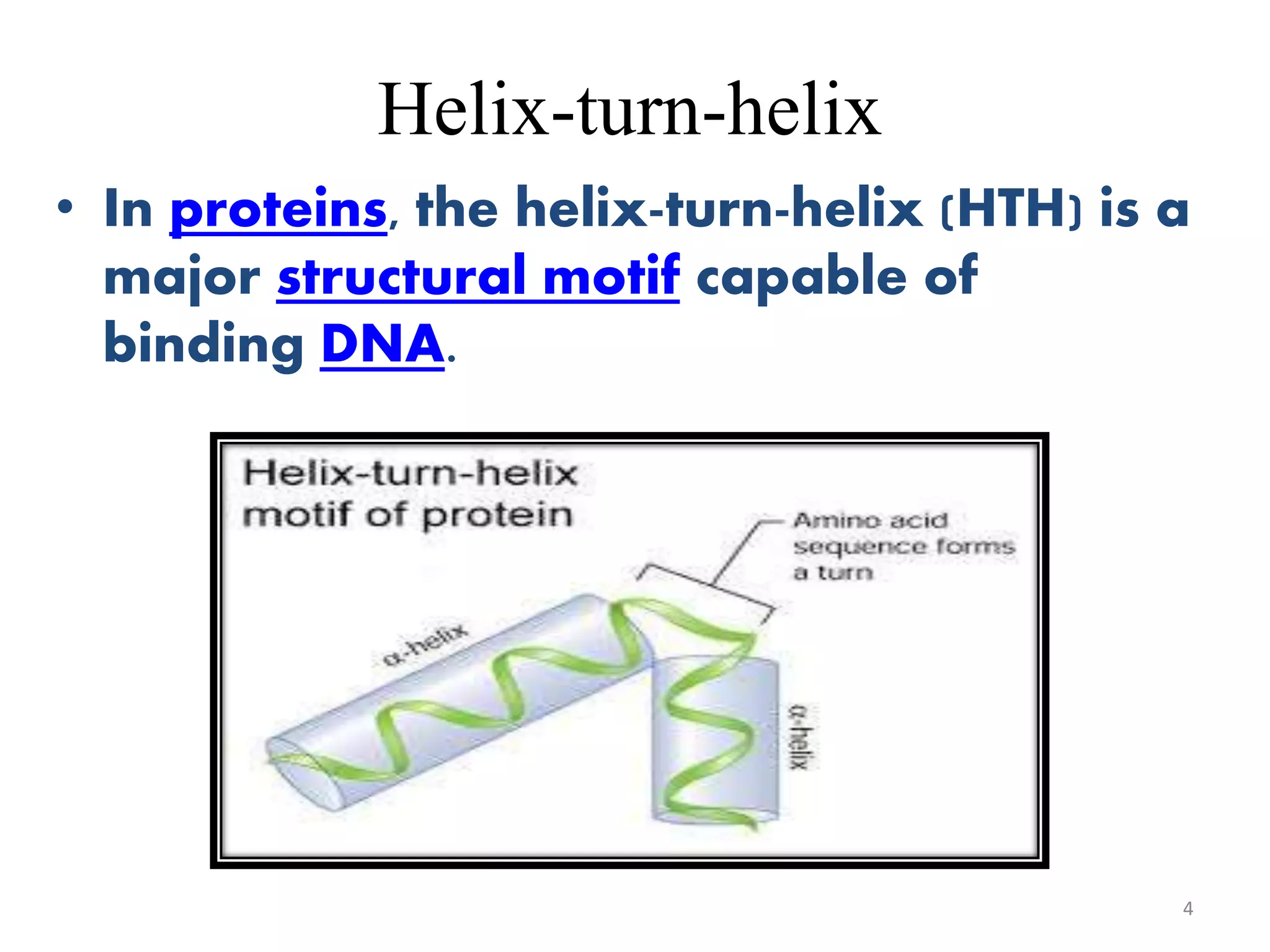
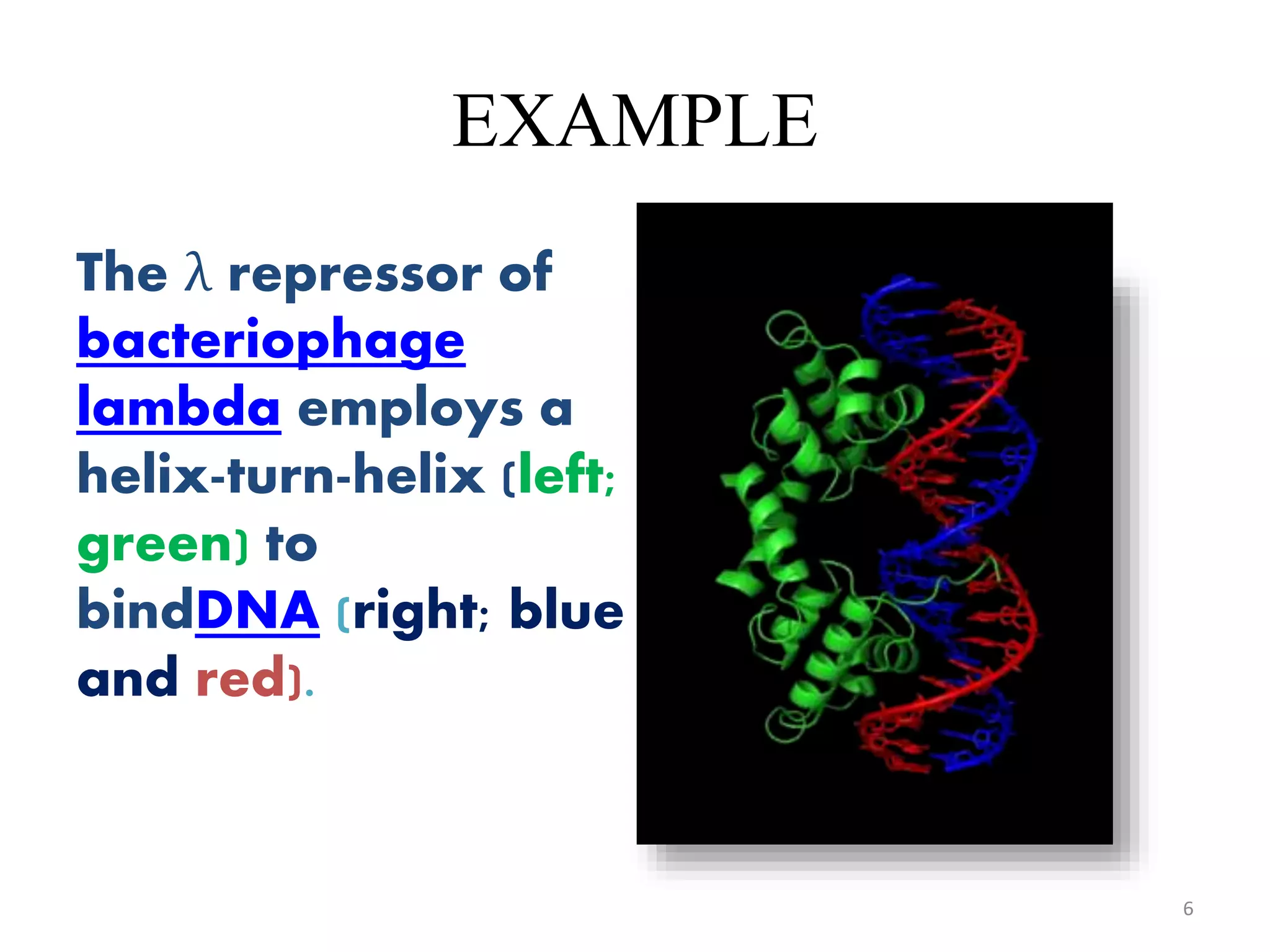
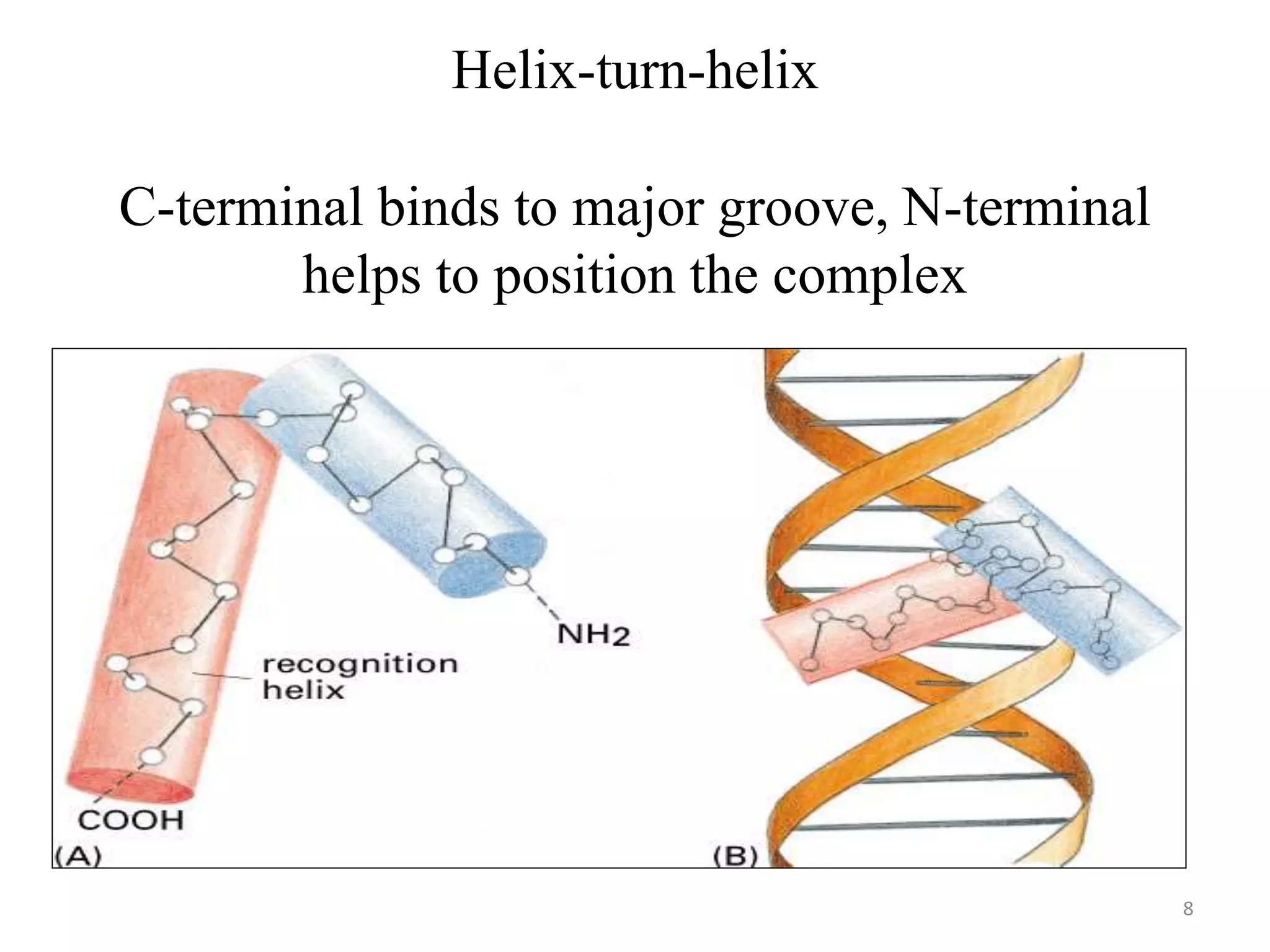
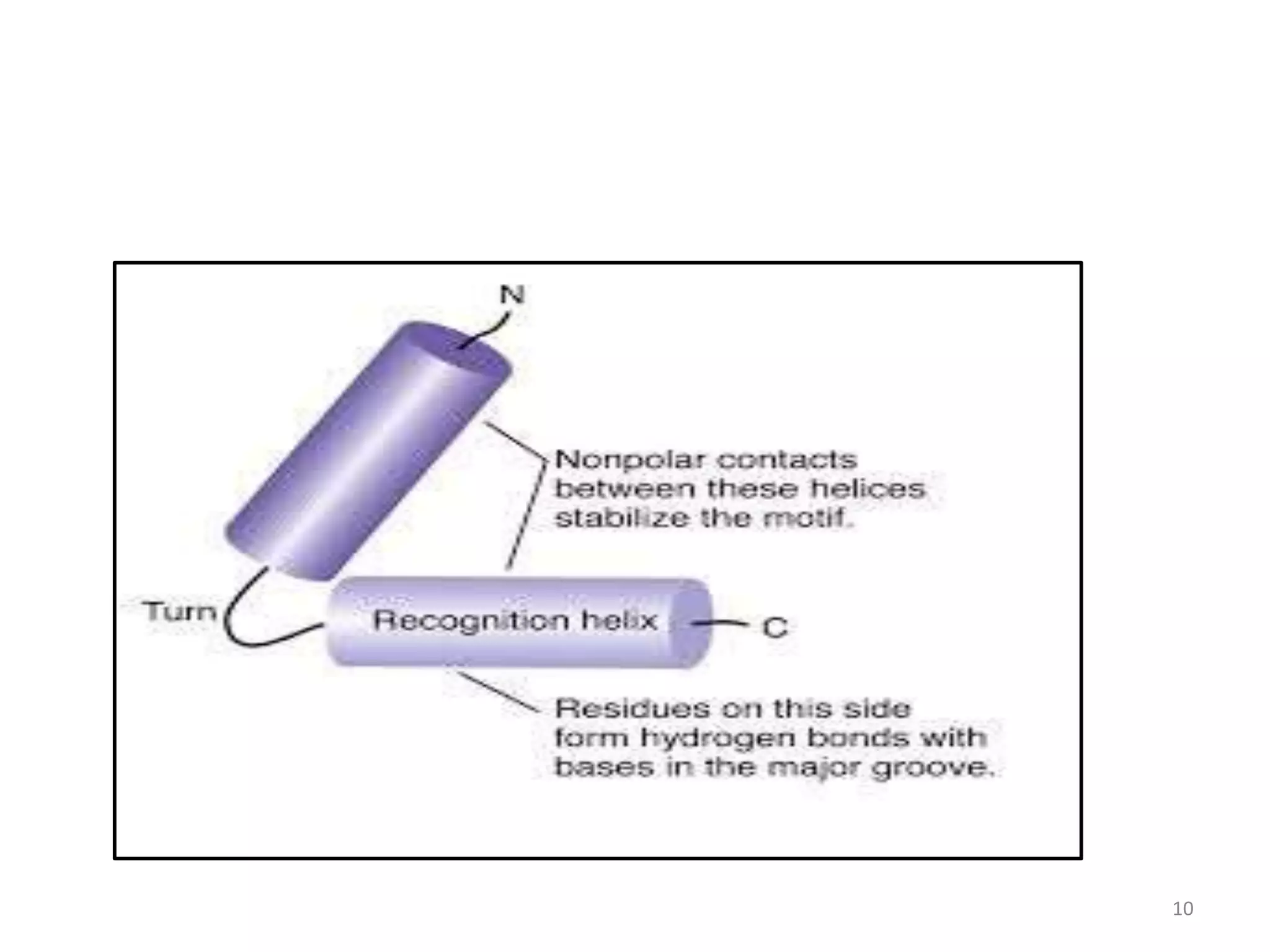
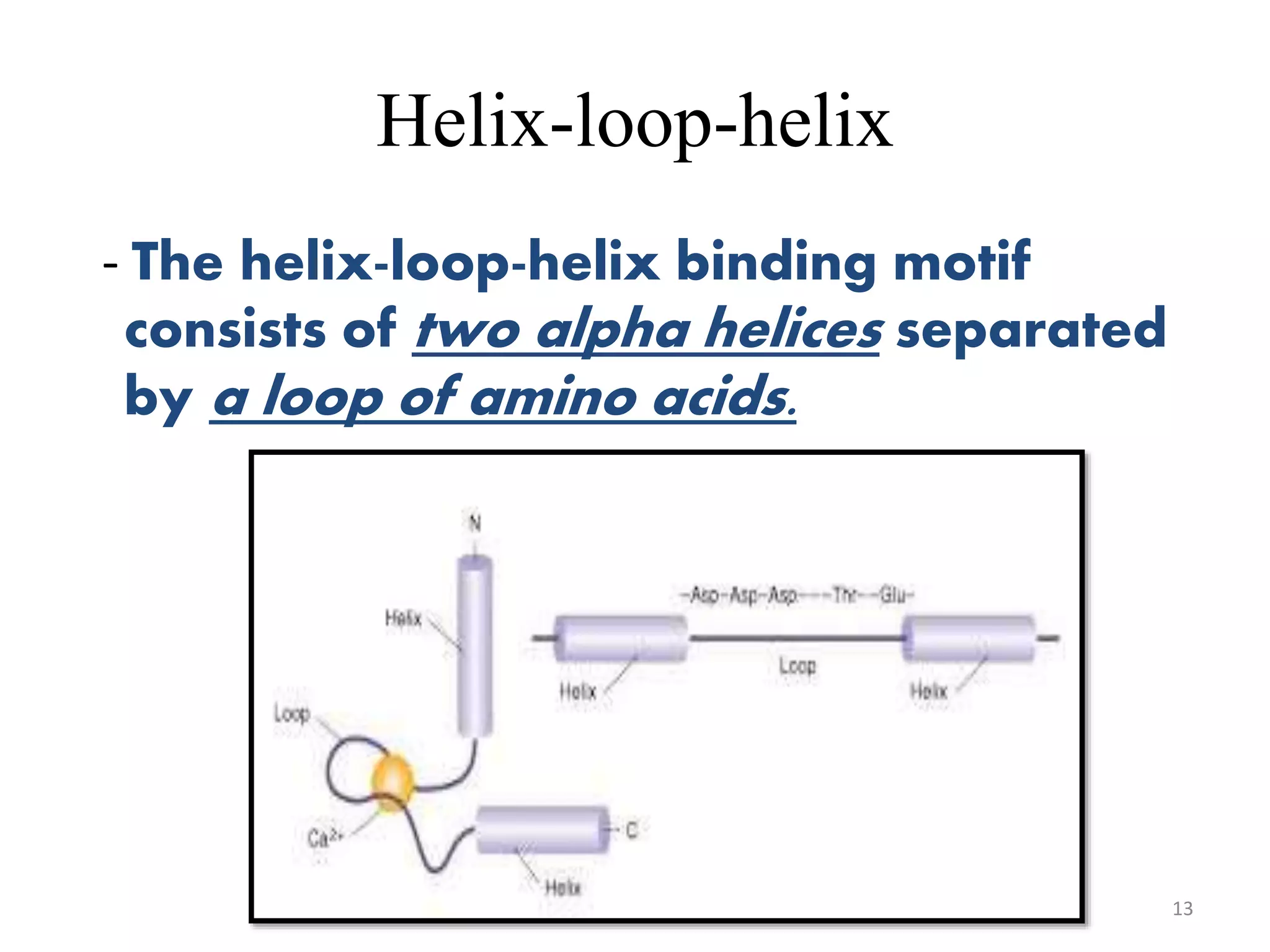
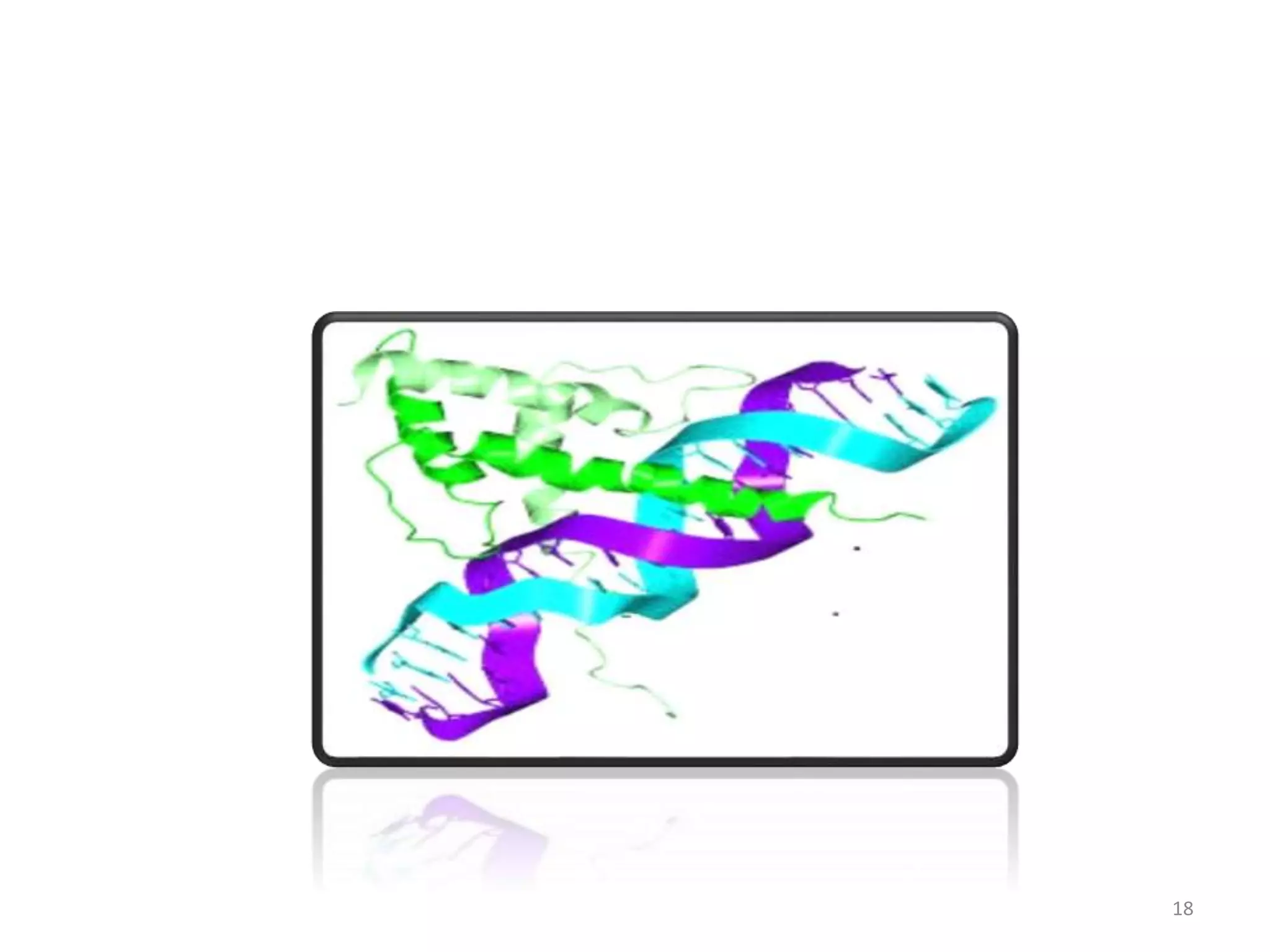
The document discusses the helix-turn-helix (HTH) and helix-loop-helix (HLH) motifs, which are critical for DNA binding in various proteins. HTH is characterized by two alpha helices that bind to the DNA's major groove and is mainly found in bacterial regulatory proteins, while HLH consists of two helices separated by a flexible loop, often found in eukaryotic transcription factors. Both motifs play significant roles in gene regulation and development, including implications in cancer through certain transcription factors.