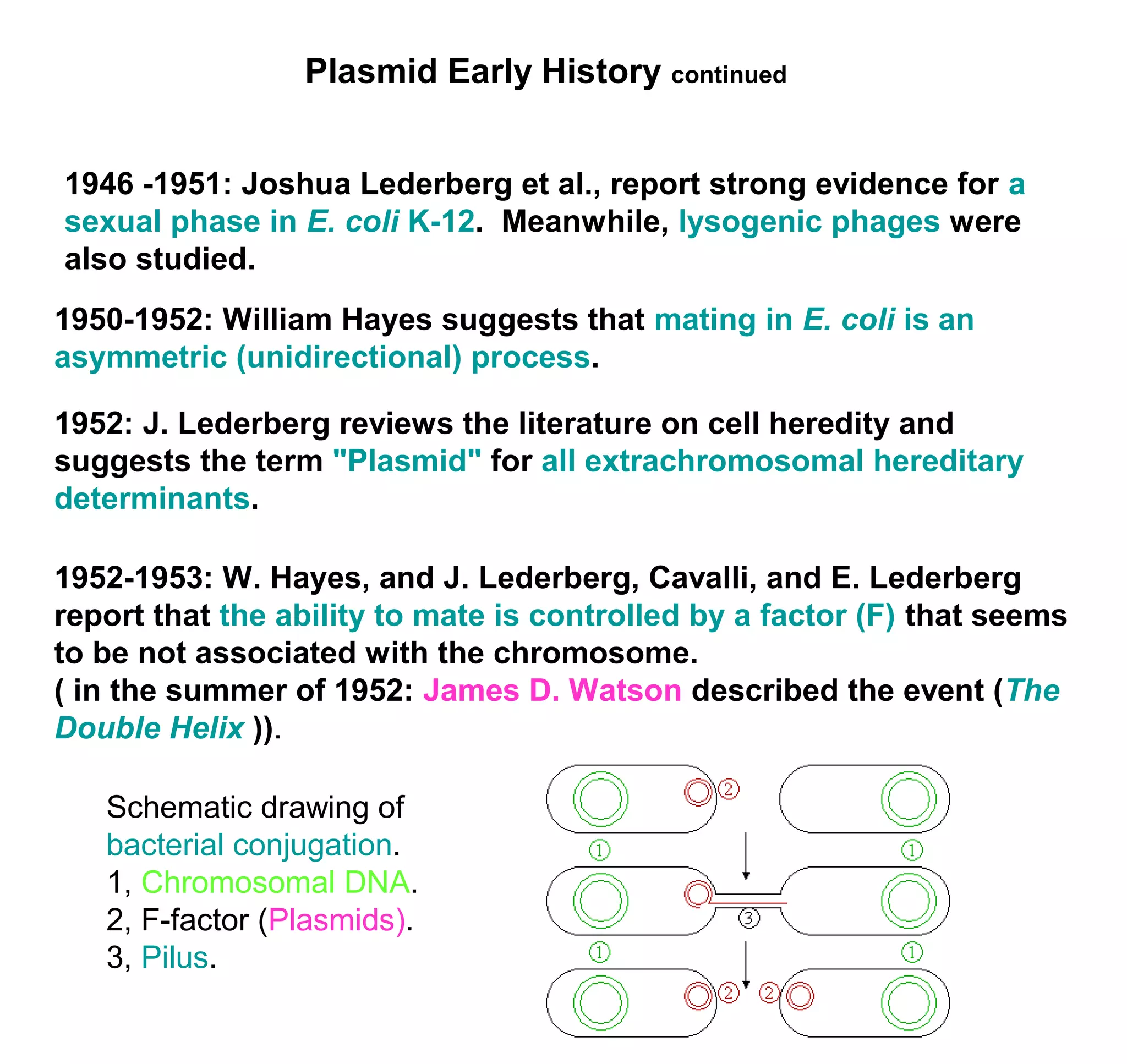
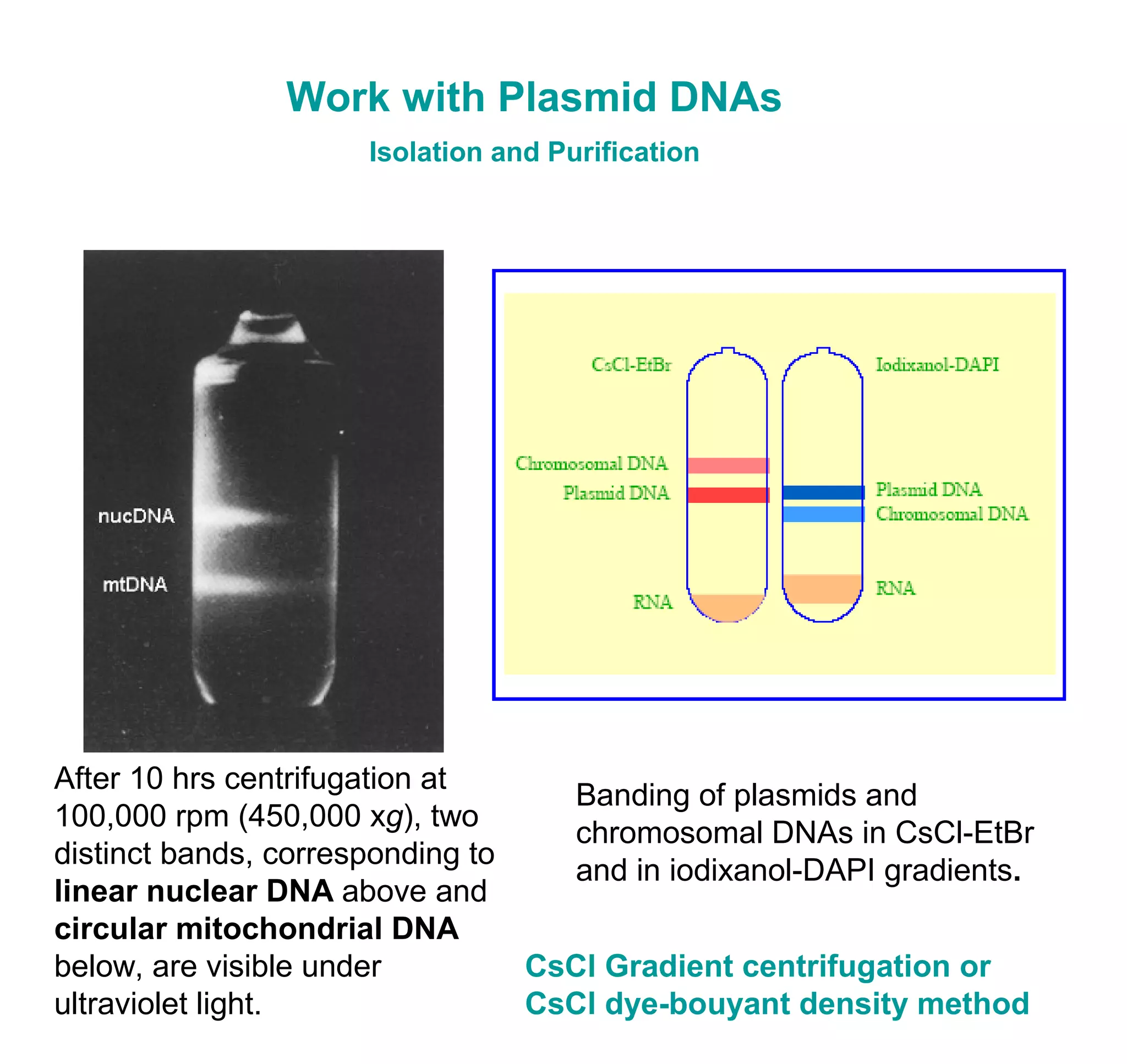
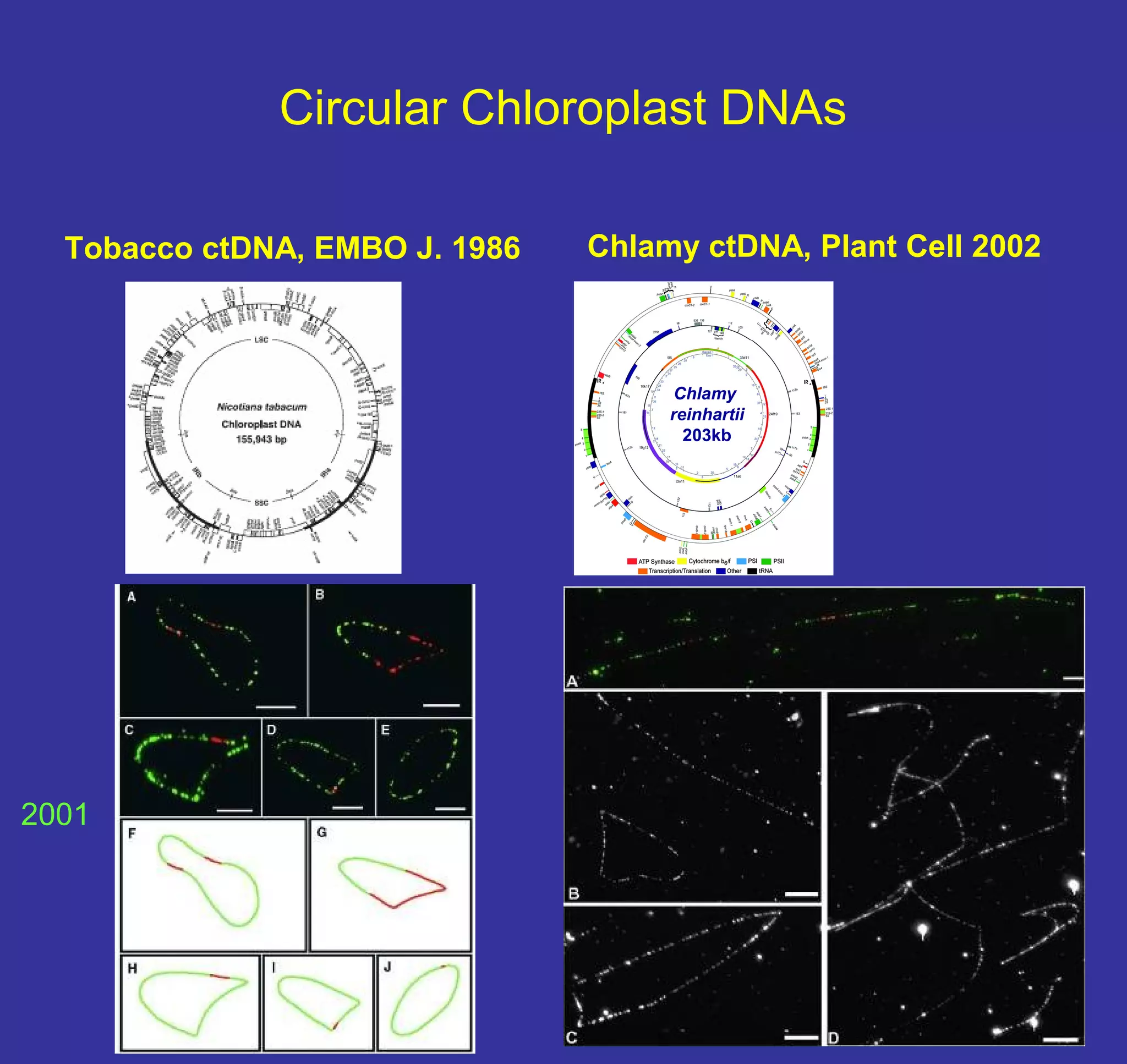
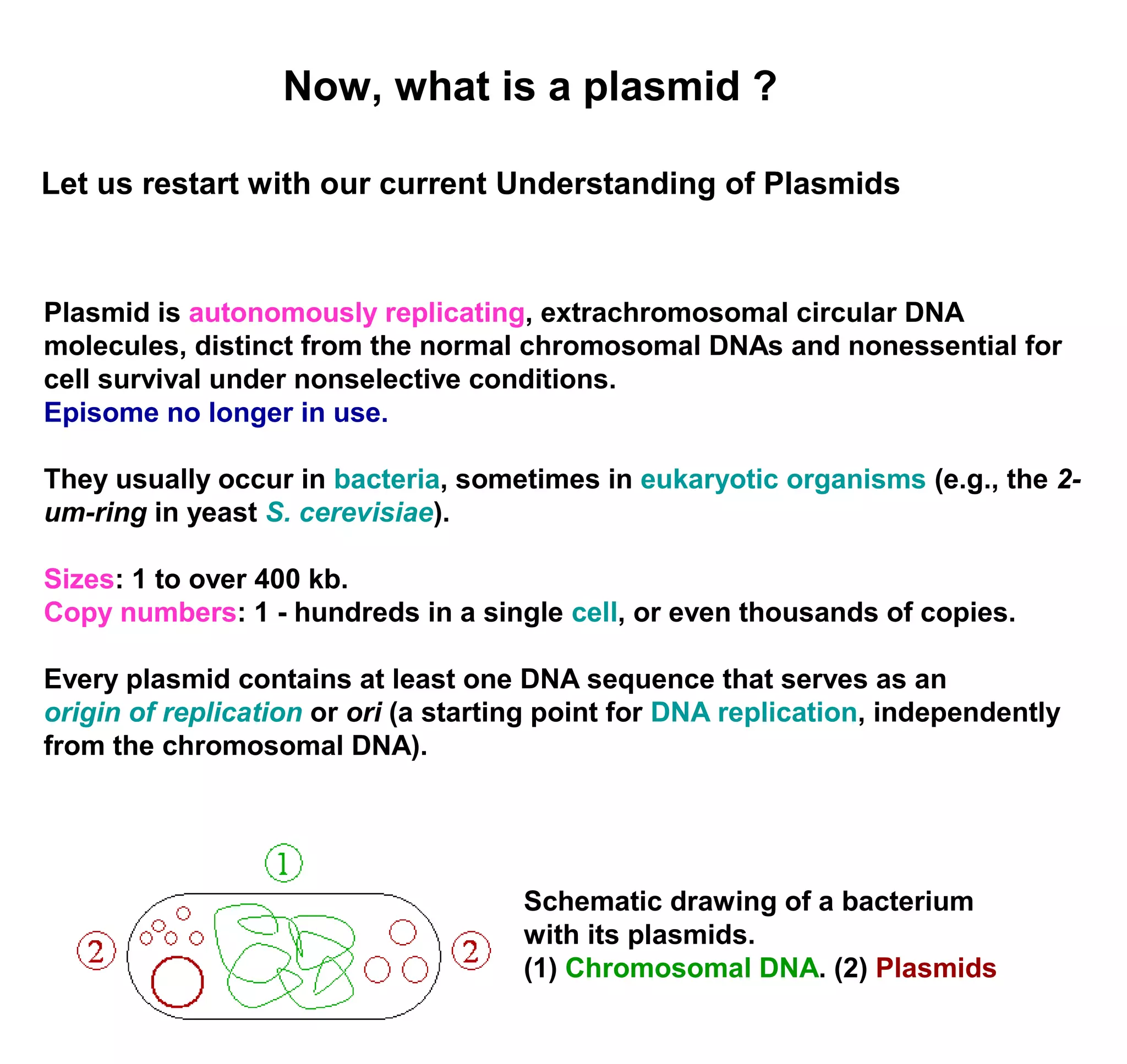
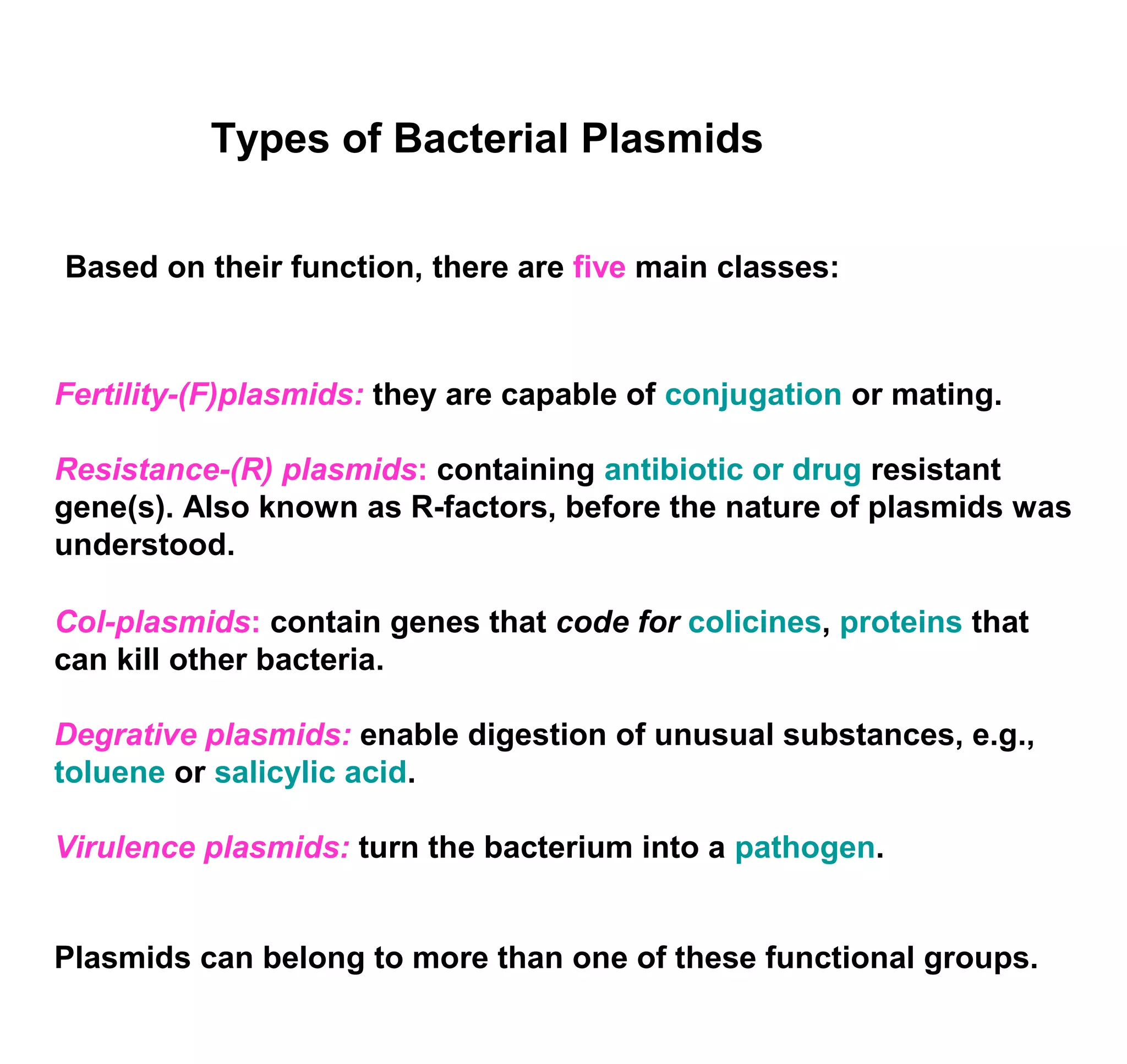
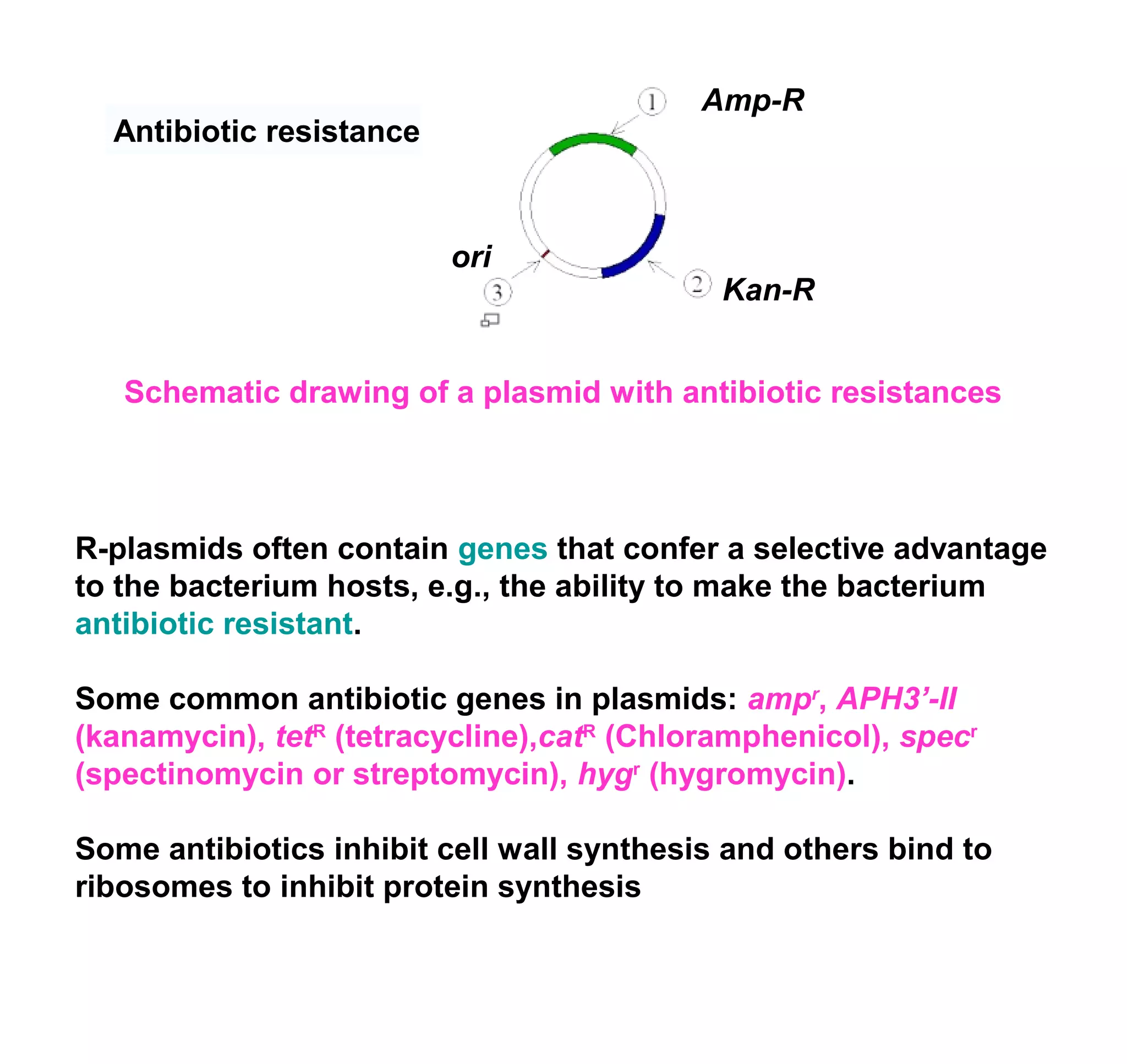
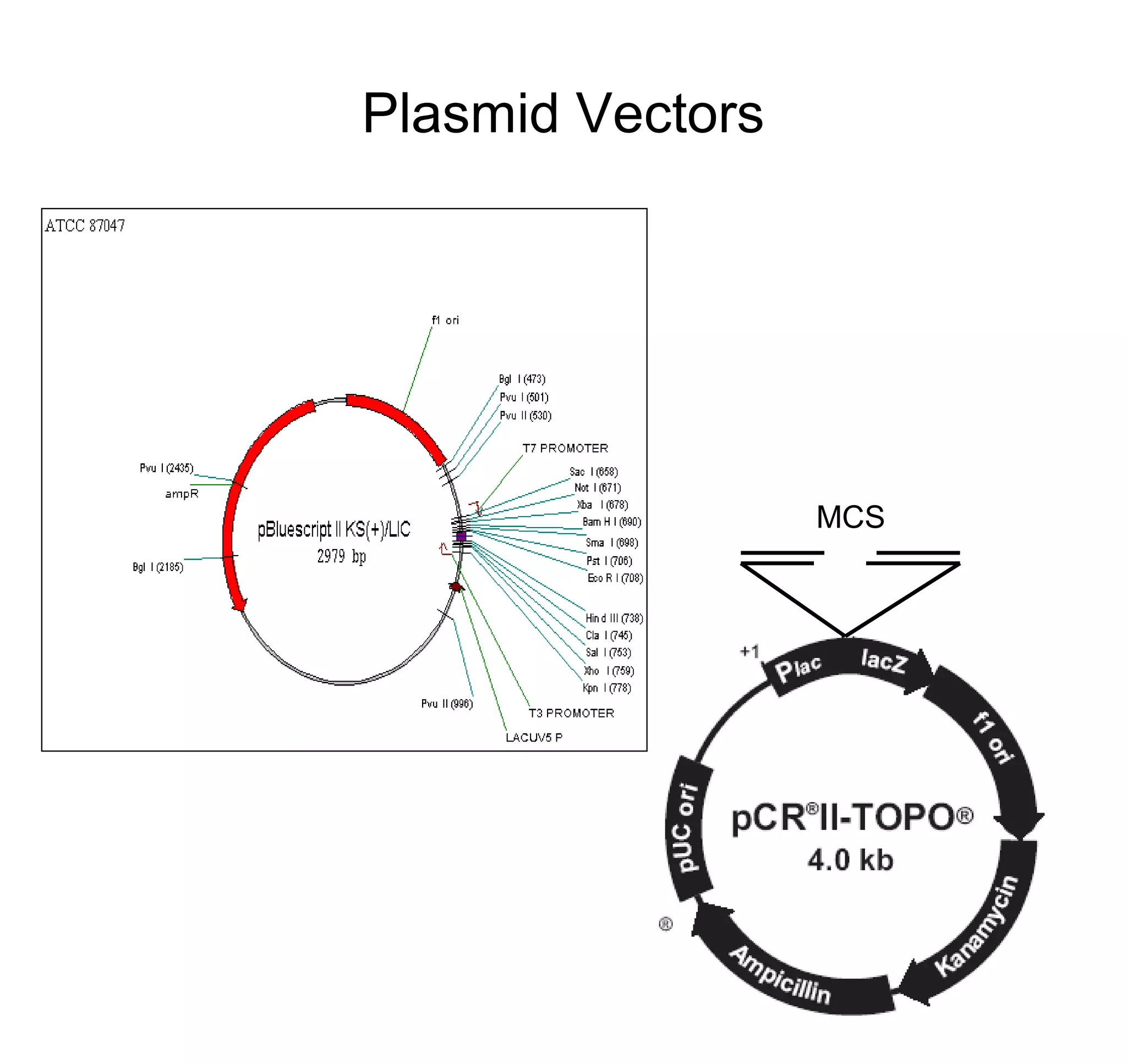
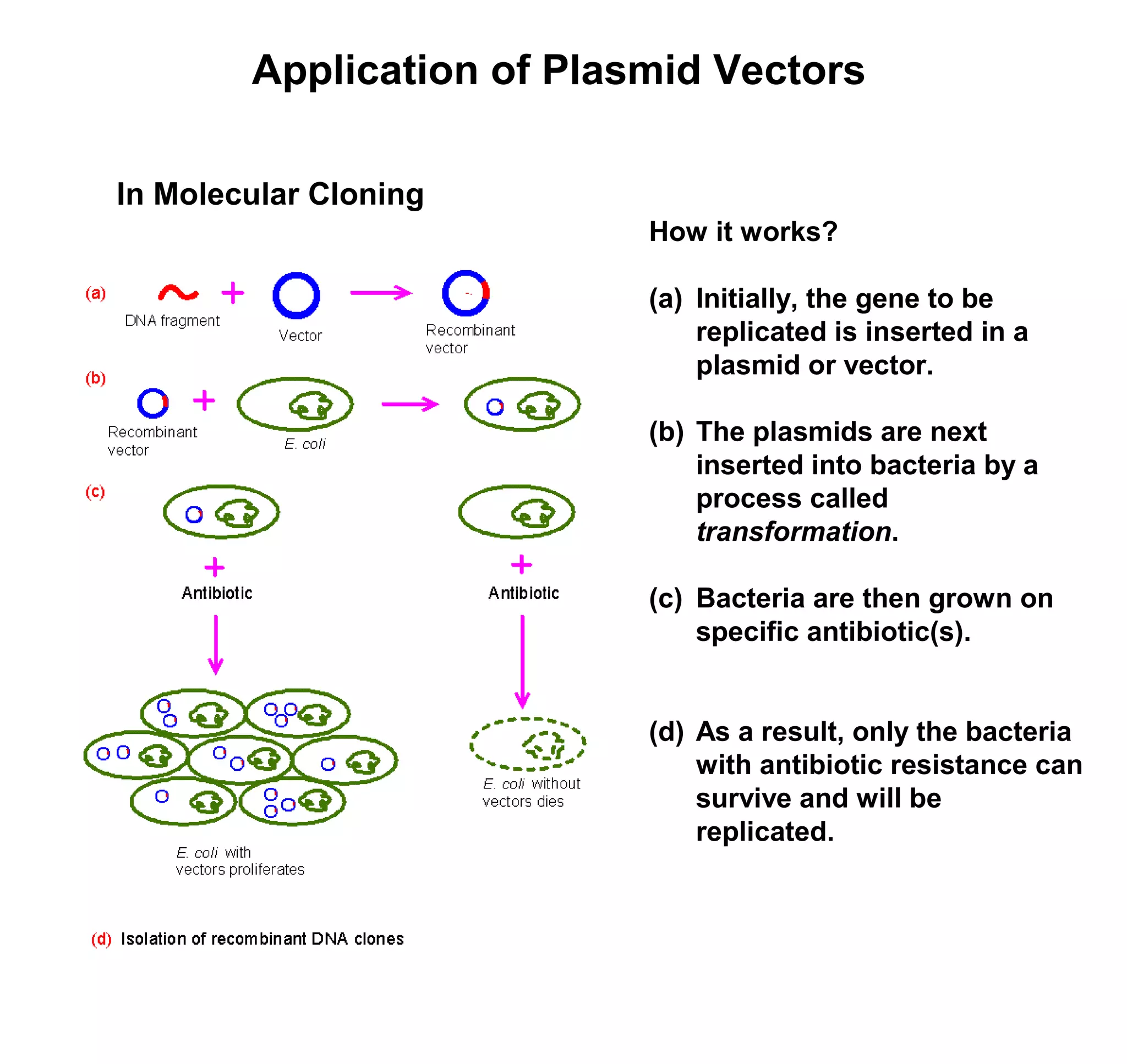
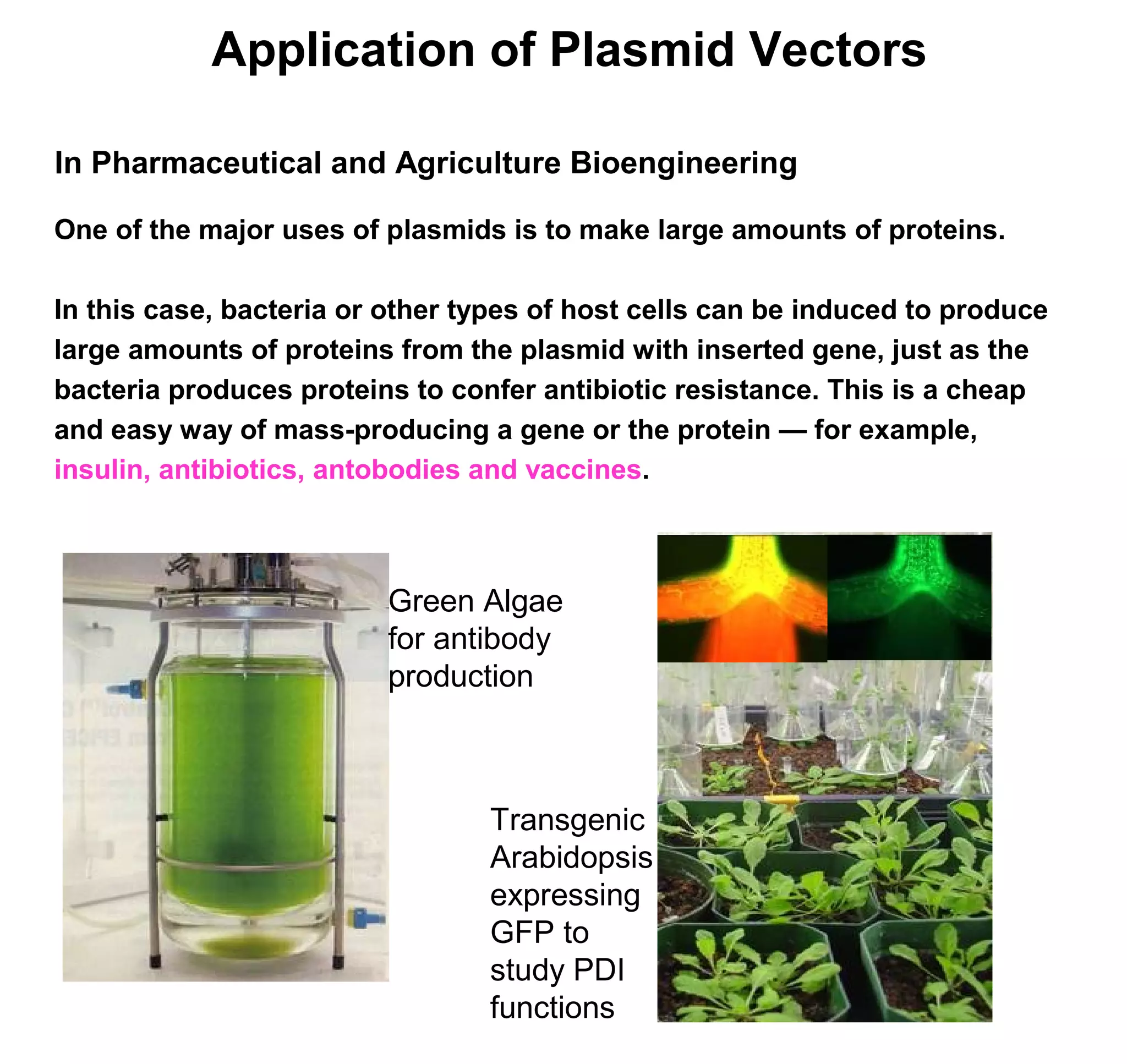
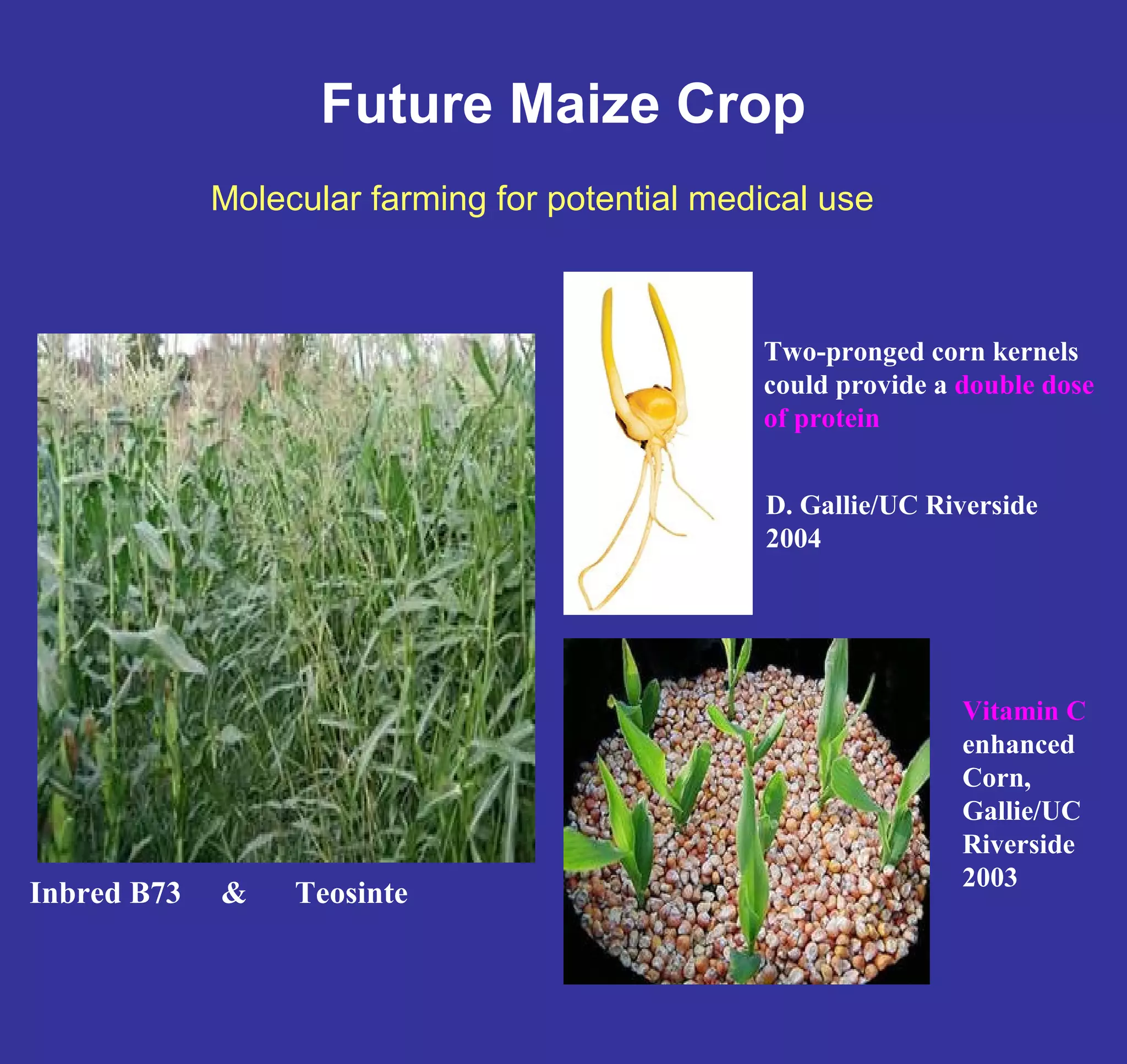
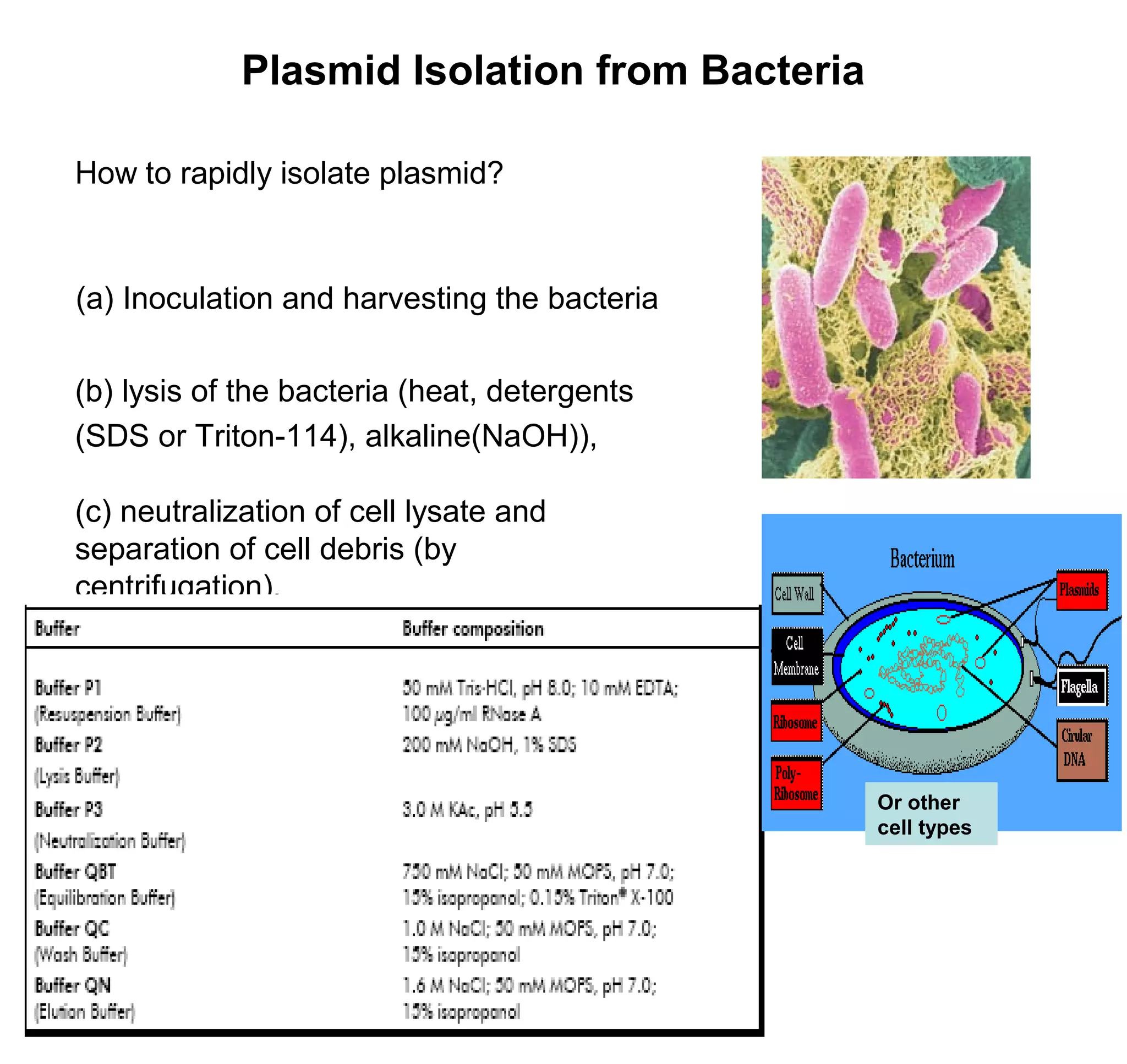
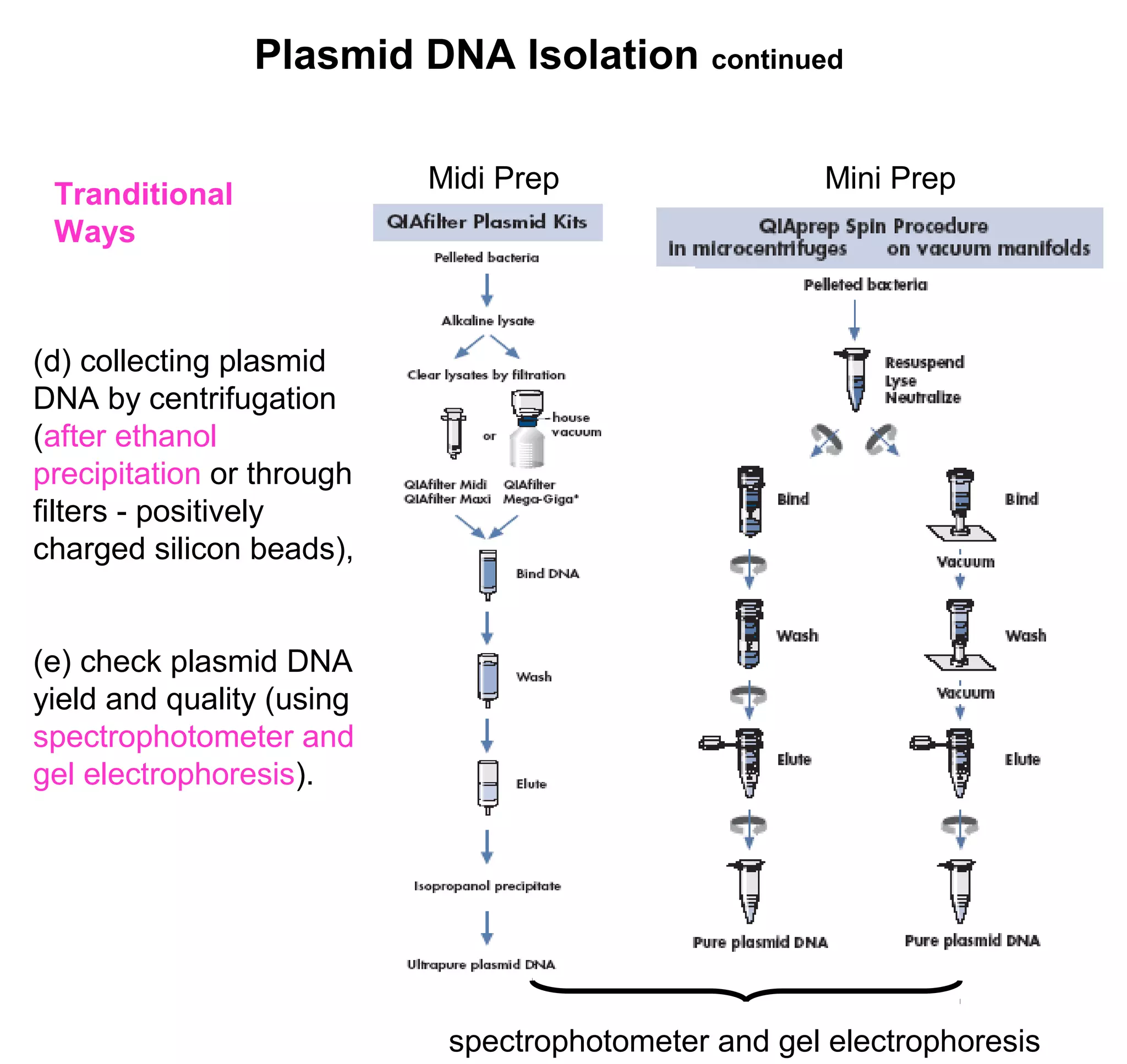
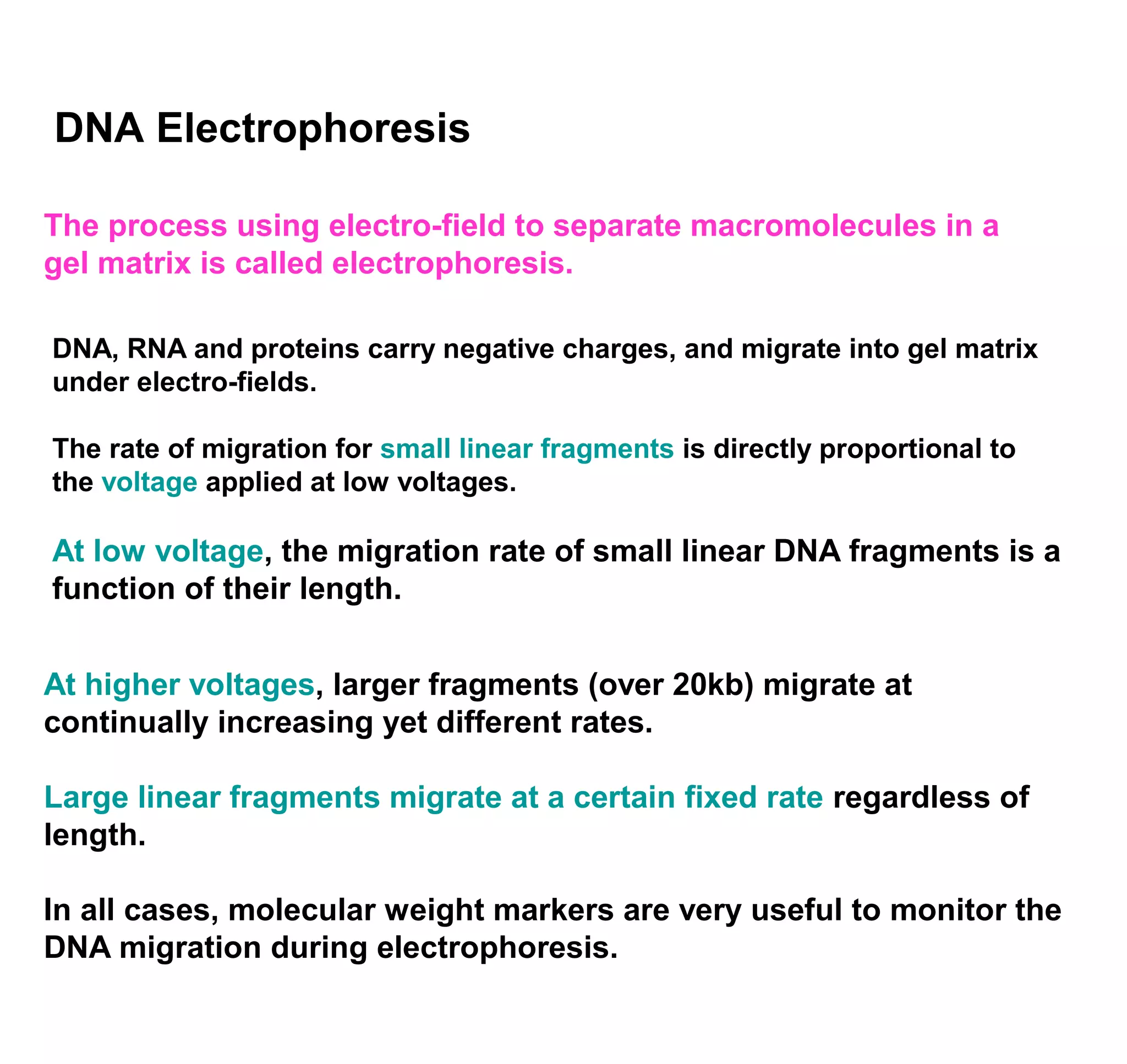
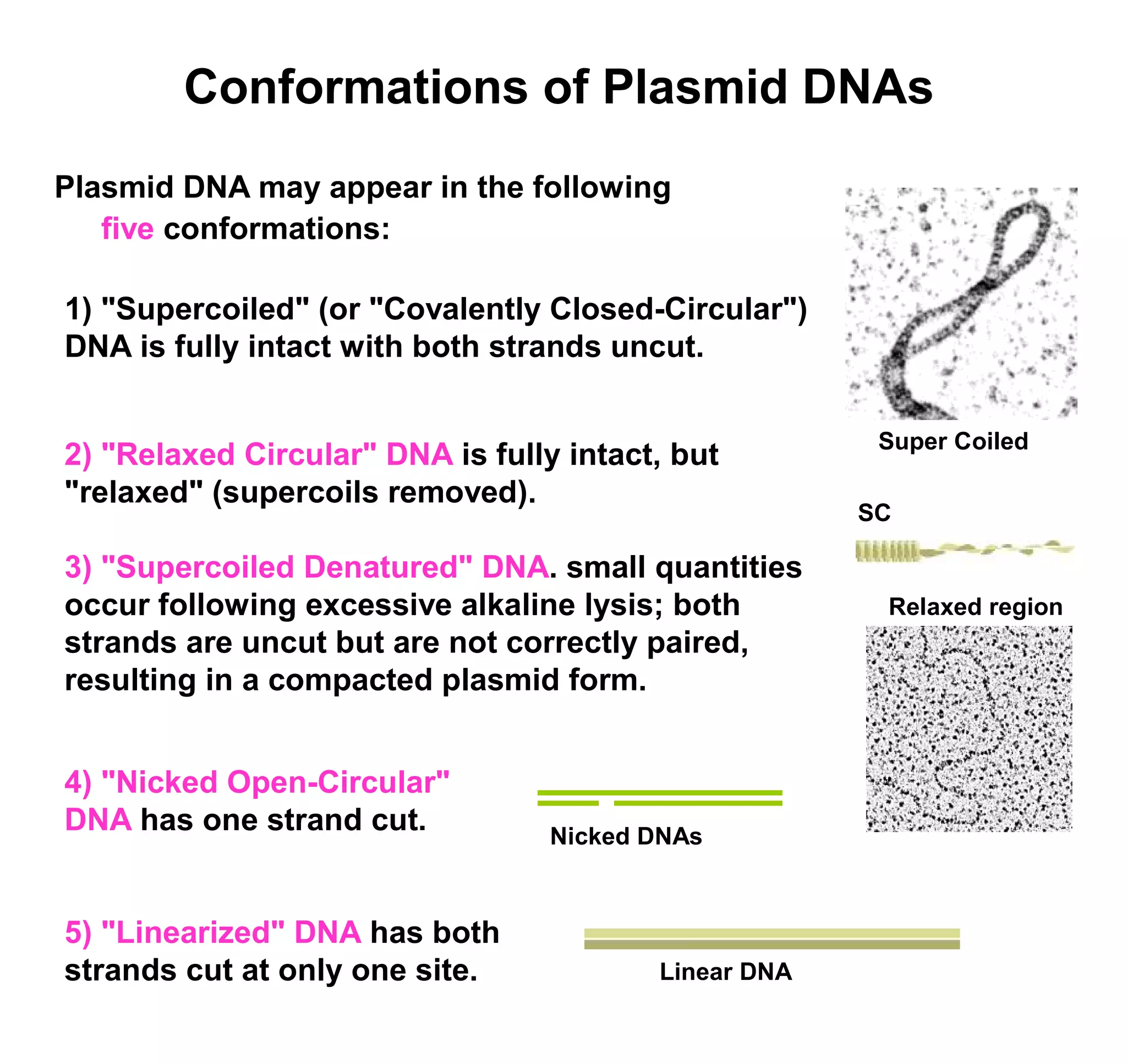
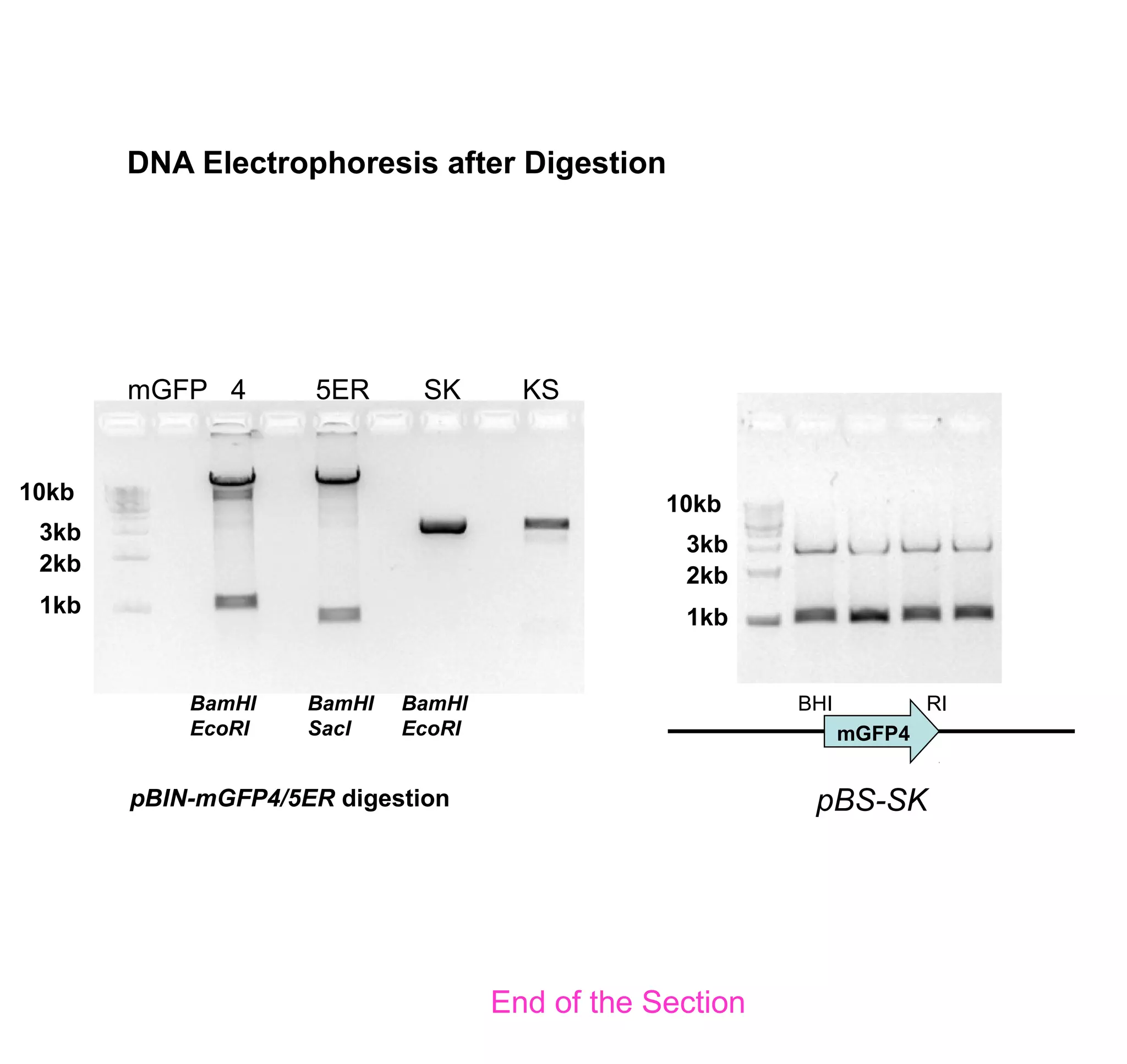
This document discusses the history and properties of plasmids. Plasmids are extrachromosomal DNA molecules that are able to replicate independently of a cell's chromosomal DNA. They were first observed in bacteria in the early 1950s and play important roles in processes like antibiotic resistance and virulence. The document outlines the early studies that helped characterize plasmids and describes some of their key properties, such as their circular structure and ability to exist in different conformations. It also discusses how plasmids are used as cloning vectors to amplify genes and produce proteins for applications in research, medicine, and agriculture.