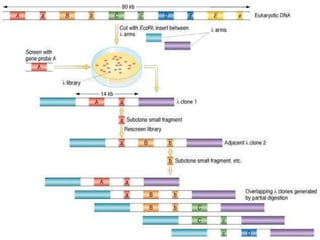
Chromosome walking is a method used to isolate and clone a particular gene or allele through positional cloning. It involves using overlapping clones that contain DNA fragments near the target gene to "walk" through the chromosome until reaching the gene. Each successive clone is tested to map its precise location until eventually reaching the target gene. Chromosome walking was developed in the early 1980s and can be used to analyze genetically transmitted diseases and find single nucleotide polymorphisms. However, it has limitations such as being a slow process and difficulty walking through repeated sequences.