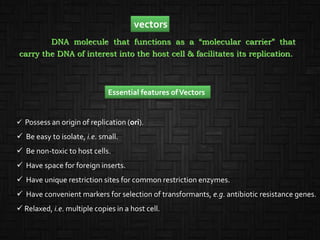
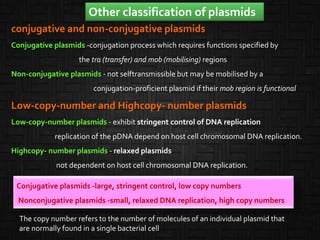
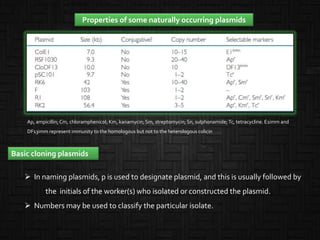
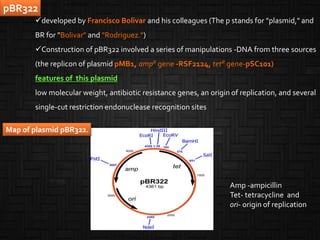
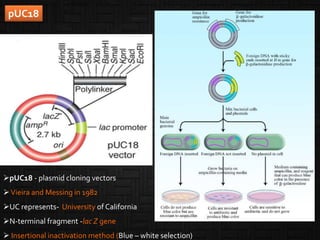
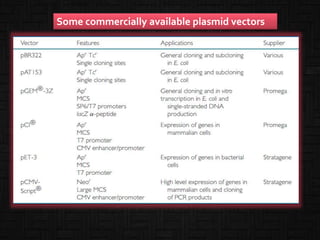
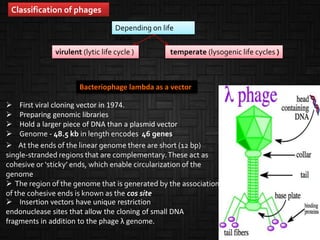
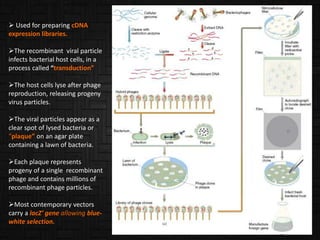
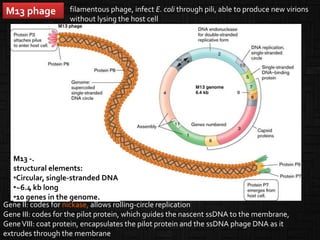
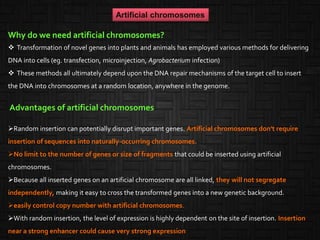
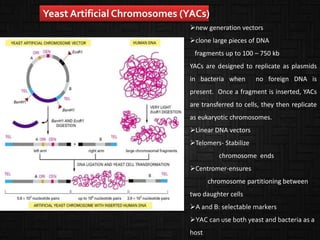
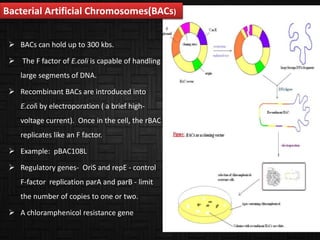
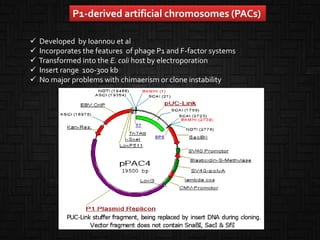
This document discusses host cells and vectors used in gene cloning. It describes various prokaryotic and eukaryotic host cells, including E. coli, yeast, and mammalian cells. It also discusses the key features and types of vectors, including plasmids, bacteriophages, cosmids, and phagemids. Plasmids are the most commonly used prokaryotic vectors and come in various types including low-copy and high-copy plasmids. Common plasmid vectors discussed include pBR322, pUC18, and commercially available vectors. Bacteriophages like lambda phage and M13 phage are also described as viral vectors.









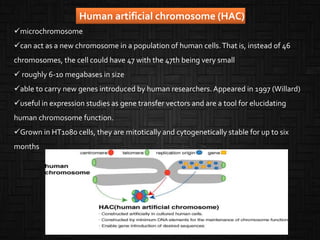
![Mammalian artificial chromosomes (MACs)
Main features for an efficient mammalian AC (MAC)
(1) a vectorial capacity up to a few megabase
(2) a manageable size for their in vitro manipulation
(3) a correct intracellular location and copy number
(4) no untoward effect on the host cell
(5)The ability to express the transgene (or transchromosome) in a physiological way
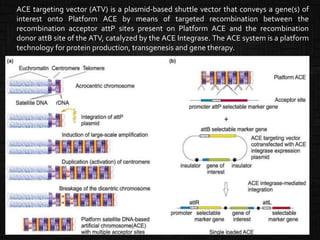
A pre-engineered platform MAC with multiple acceptor sites [Artificial
Chromosome Expression System (ACE system) ]
Capable of harboring a number of different genes
Large carrying capacity
Represents a non-integrating safe vector.
ACE Integrase, a lambda integrase enzyme, which has been modified to render
the integrase functionally independent of bacterial host cell factors and
capable of operating in a mammalian context.
ACE system)](https://image.slidesharecdn.com/hostcellandvectors-220109144355/85/Host-cell-and-vectors-25-320.jpg)