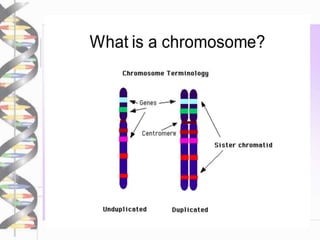
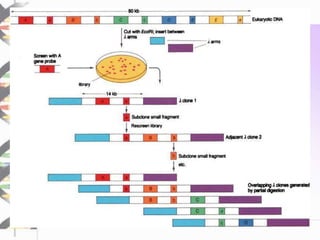
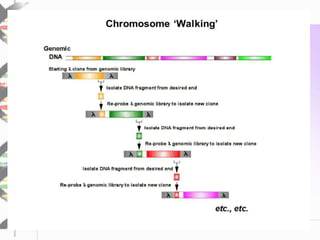
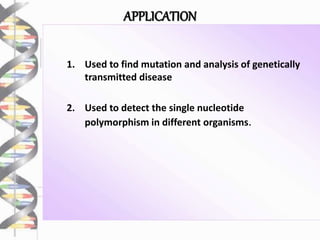
Chromosome walking is a technique used to analyze large regions of DNA by identifying overlapping clones. It involves selecting a starting clone, subcloning fragments from the ends, and using those fragments to screen a genomic library and identify neighboring clones with overlapping sequences. This process is repeated to "walk" along the chromosome. Chromosome walking was developed in the 1980s and is useful for finding mutations associated with genetic diseases and detecting single nucleotide polymorphisms, though it has limitations with repetitive sequences.