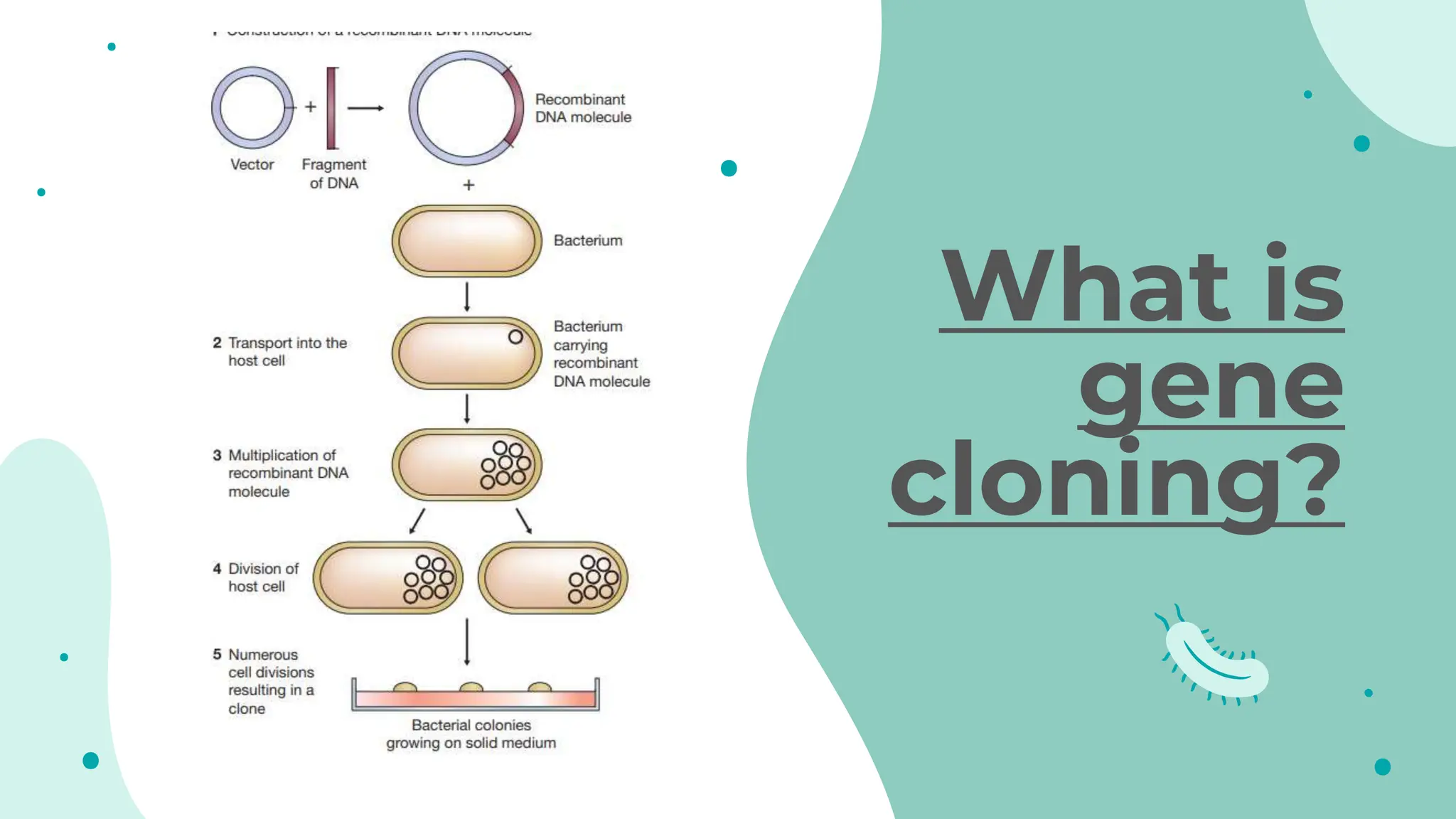
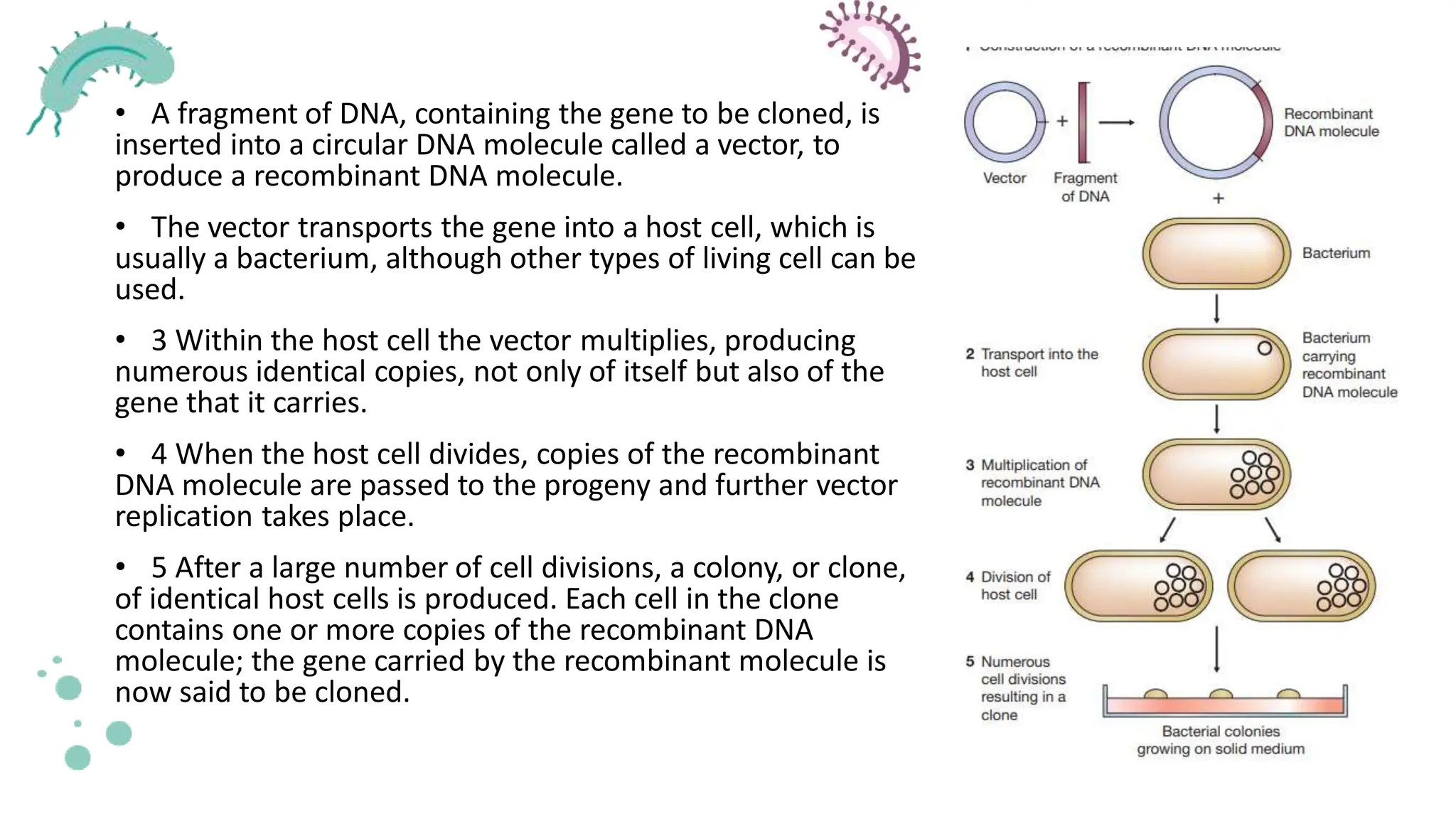
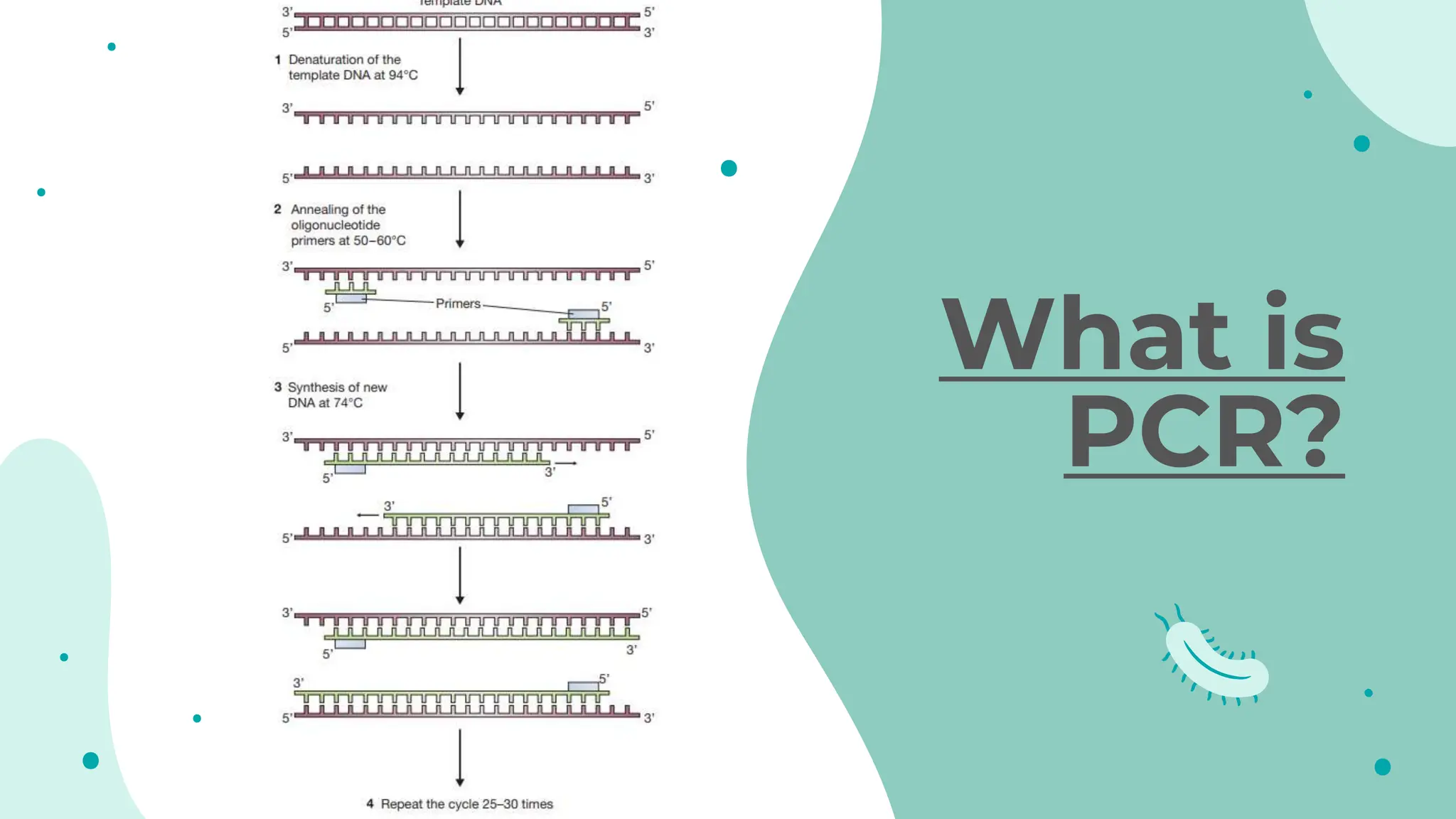
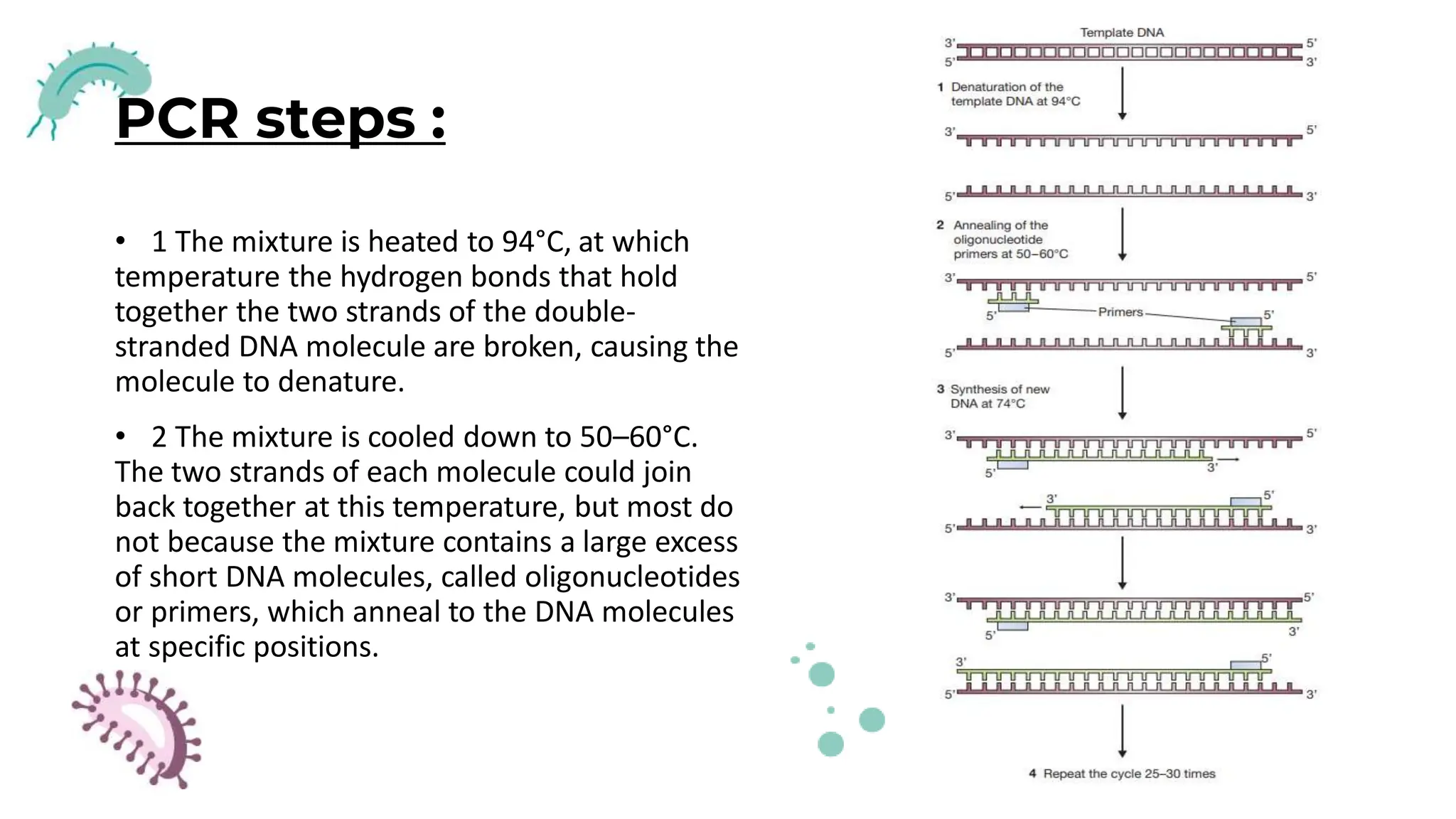
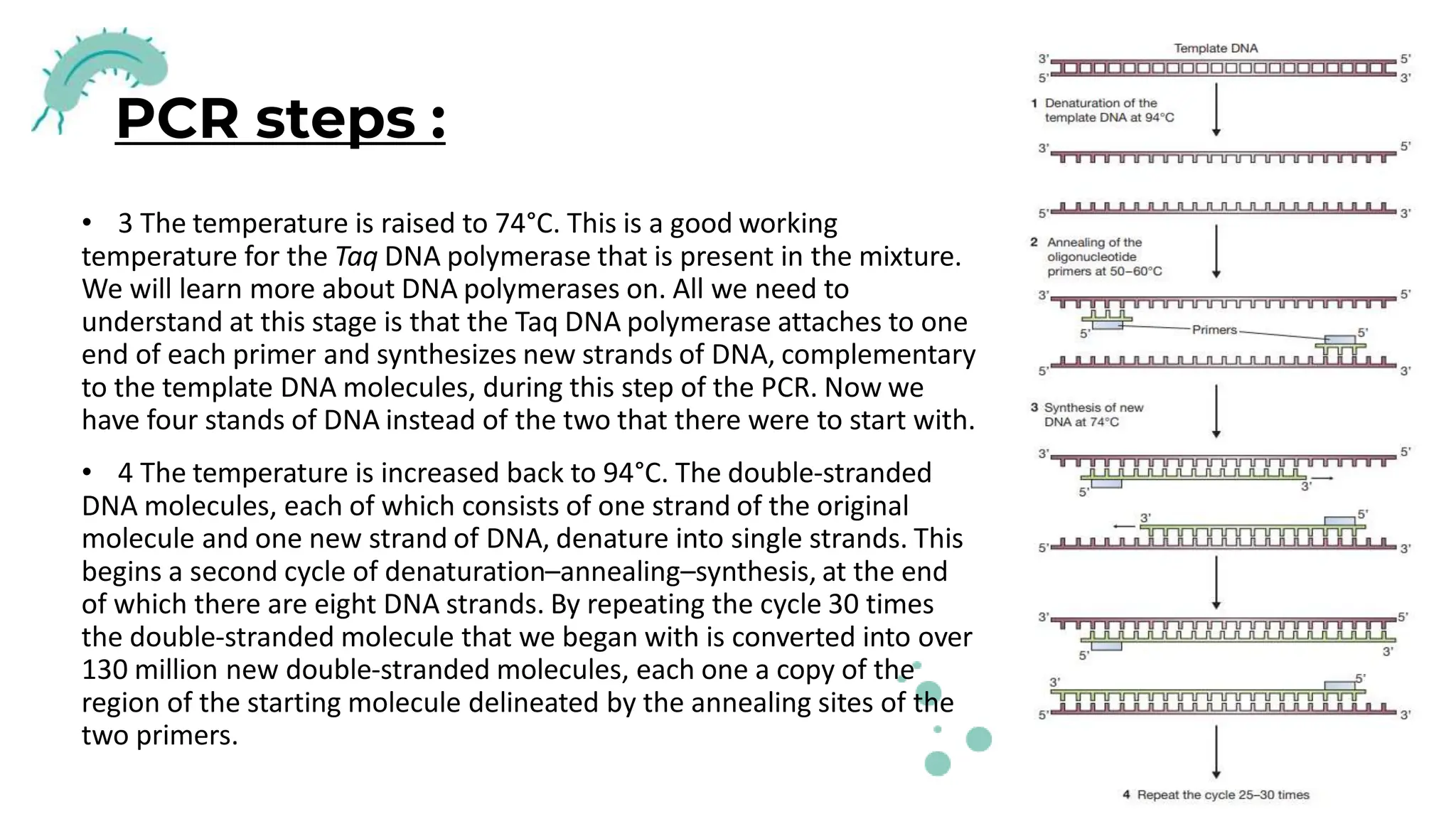
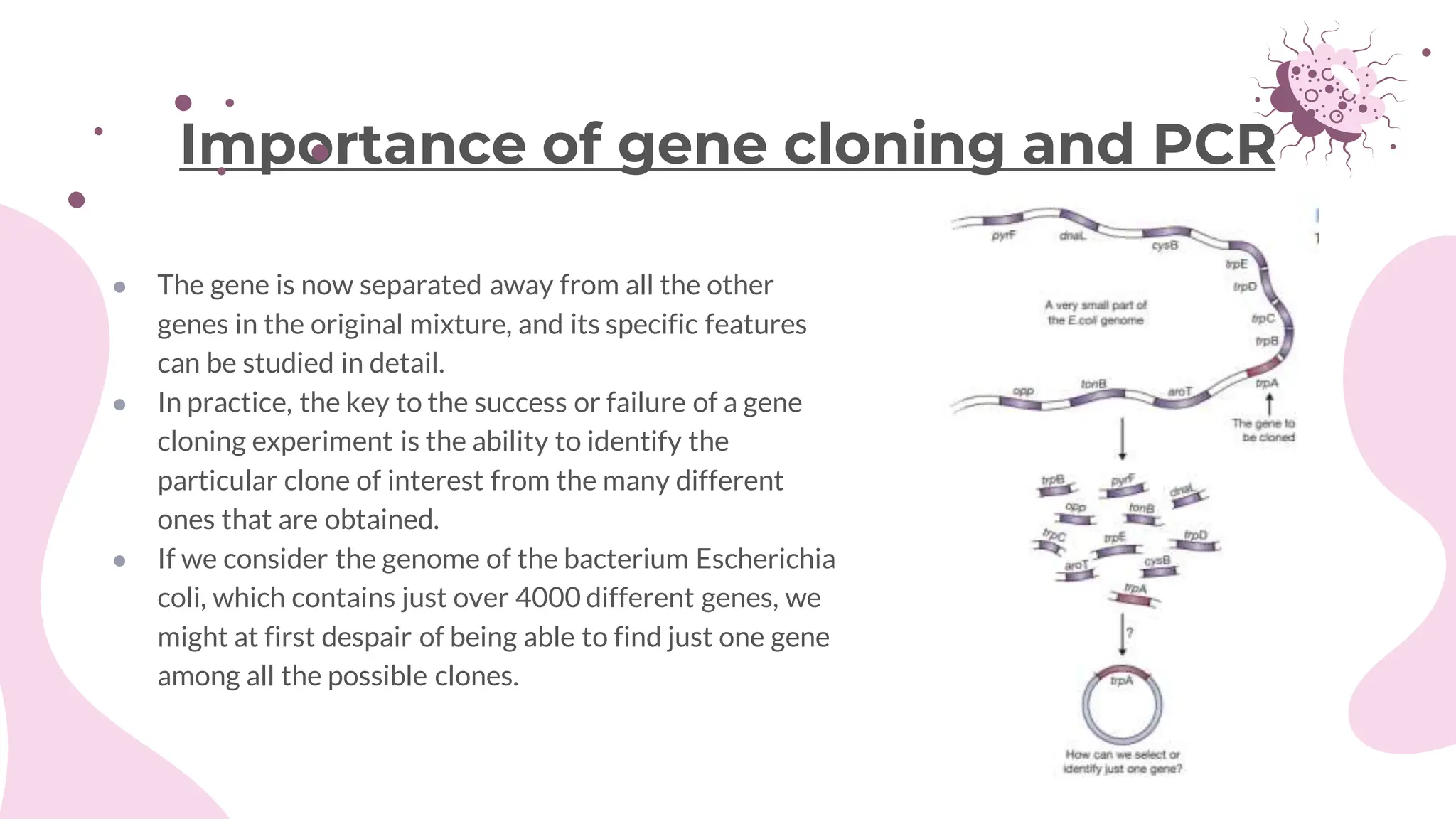
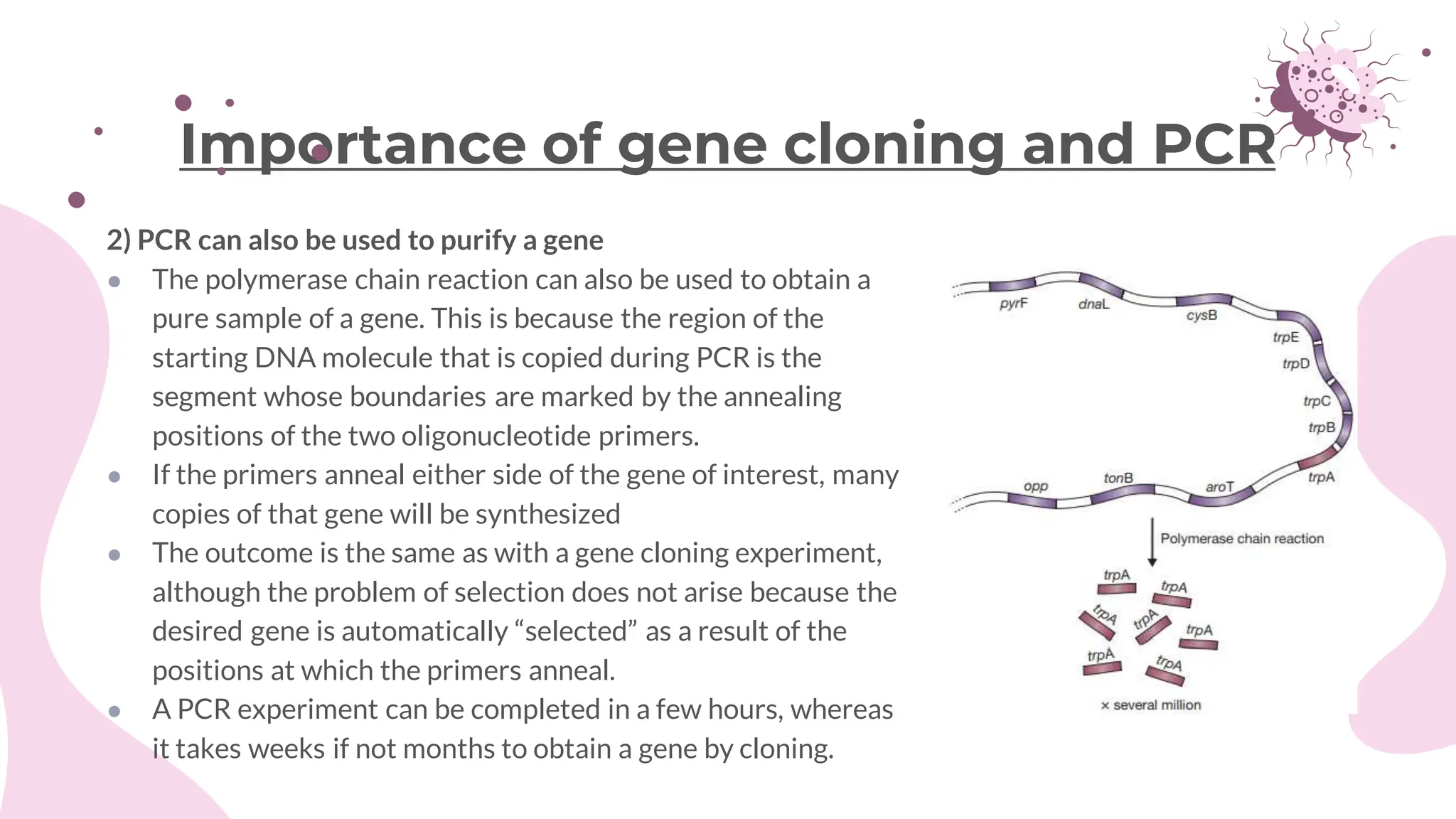
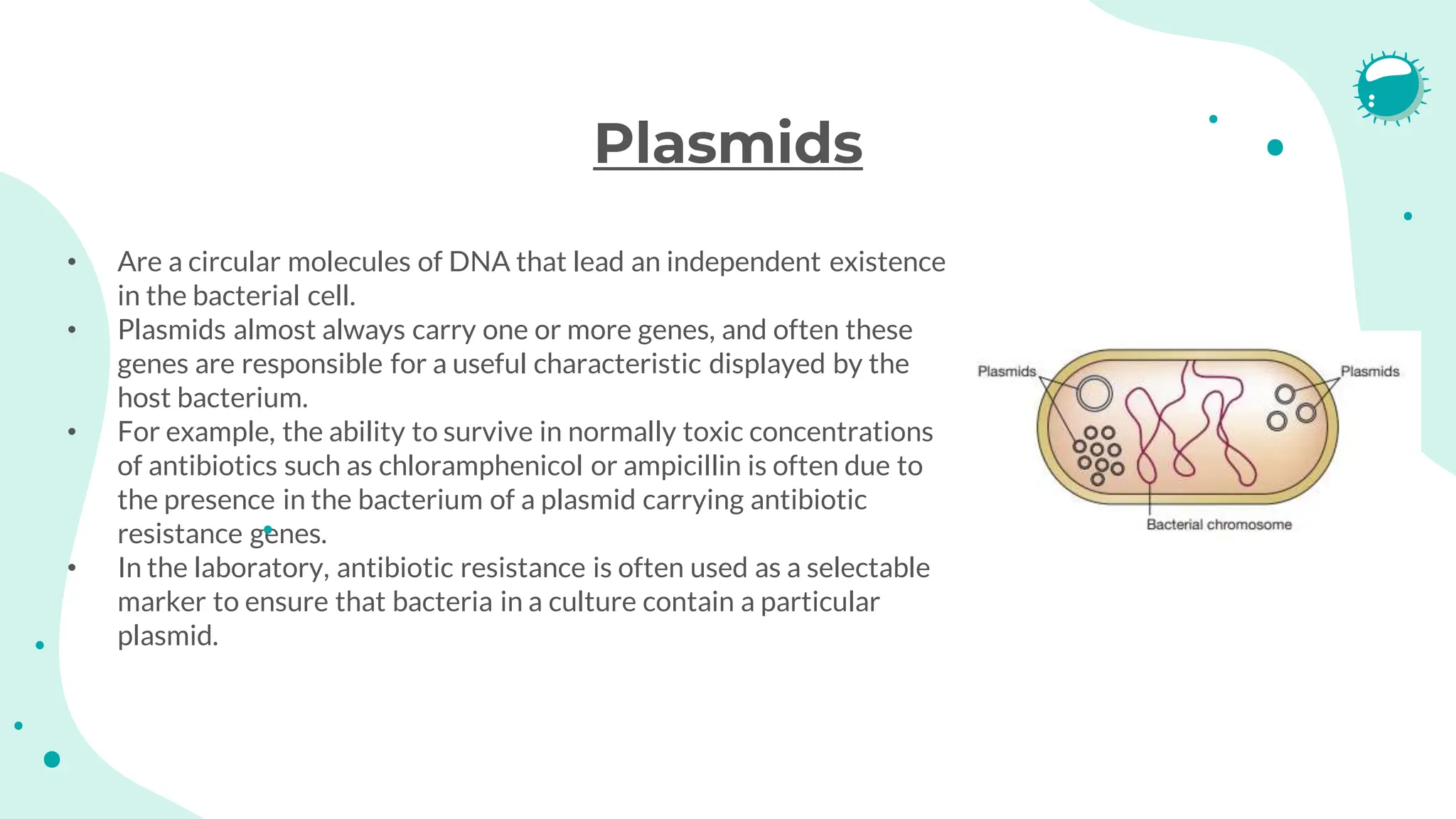
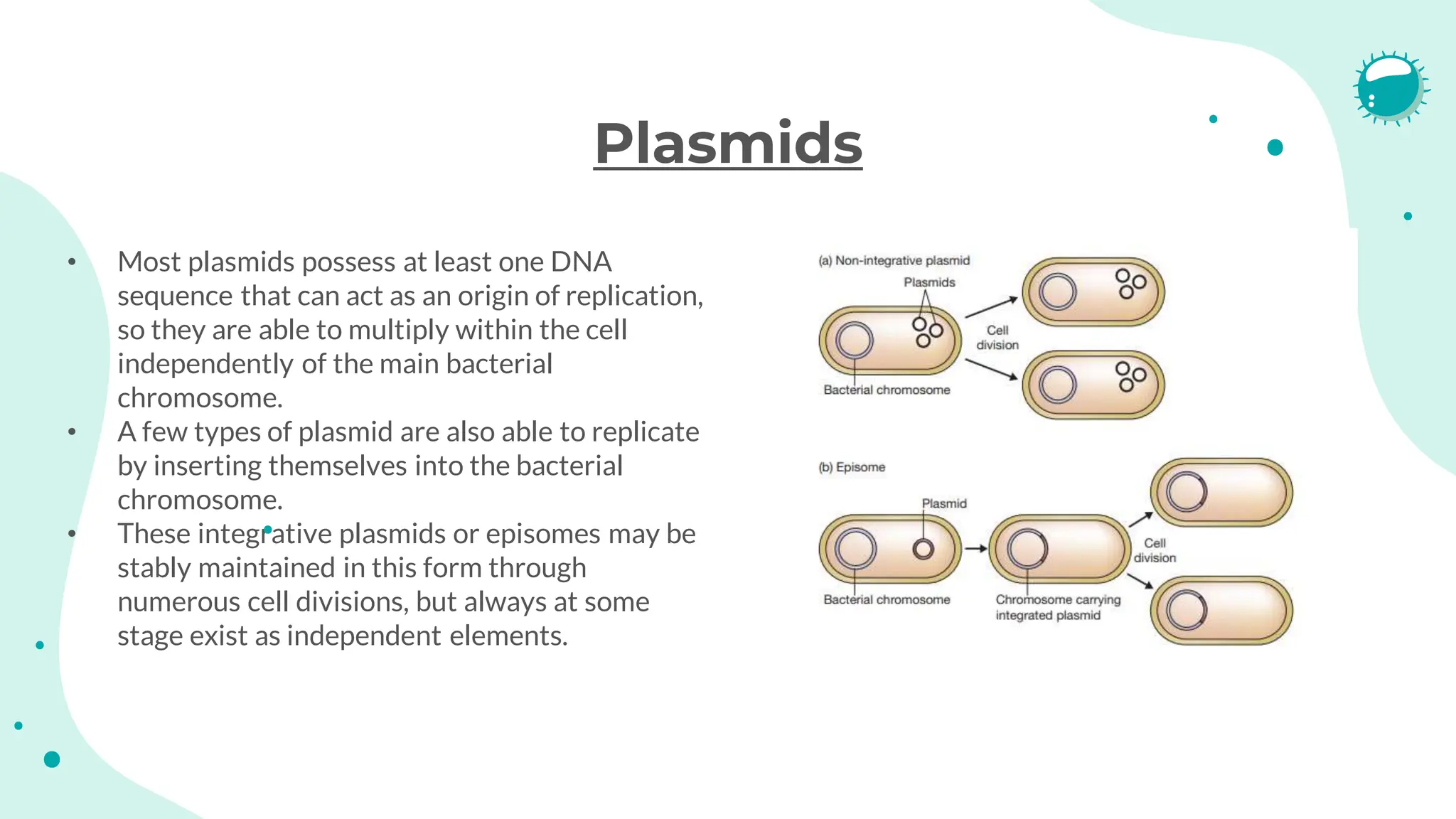
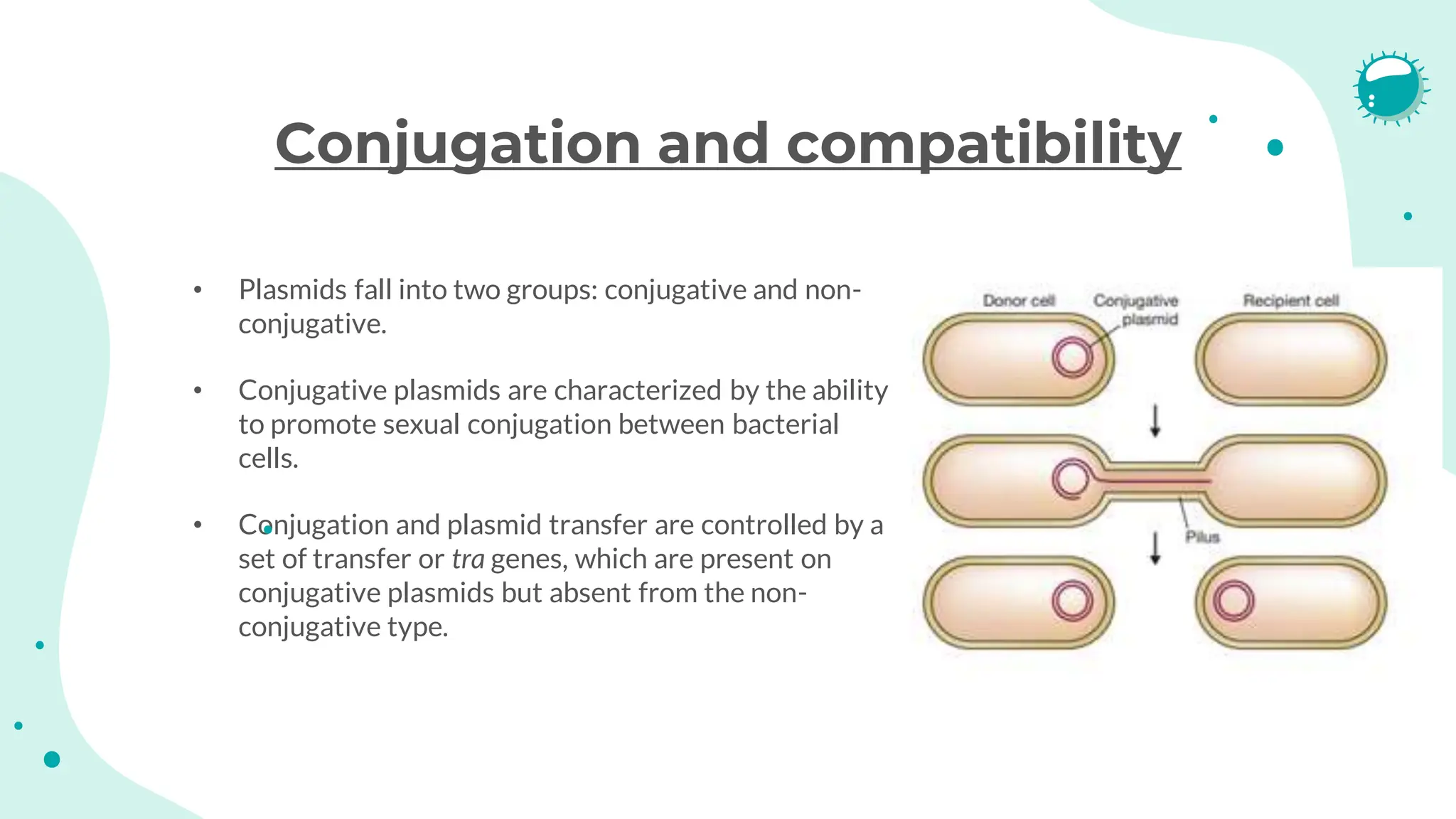
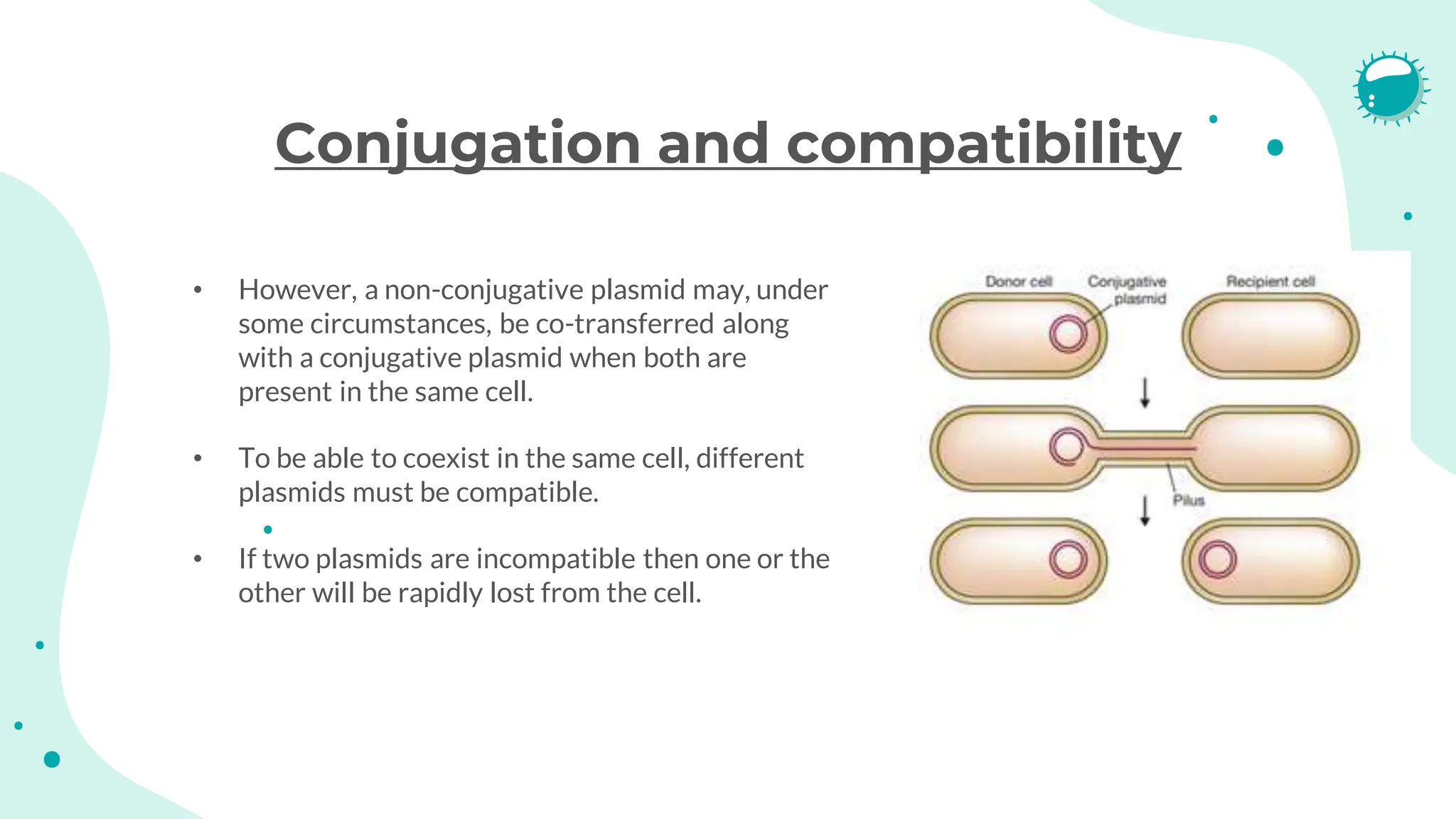
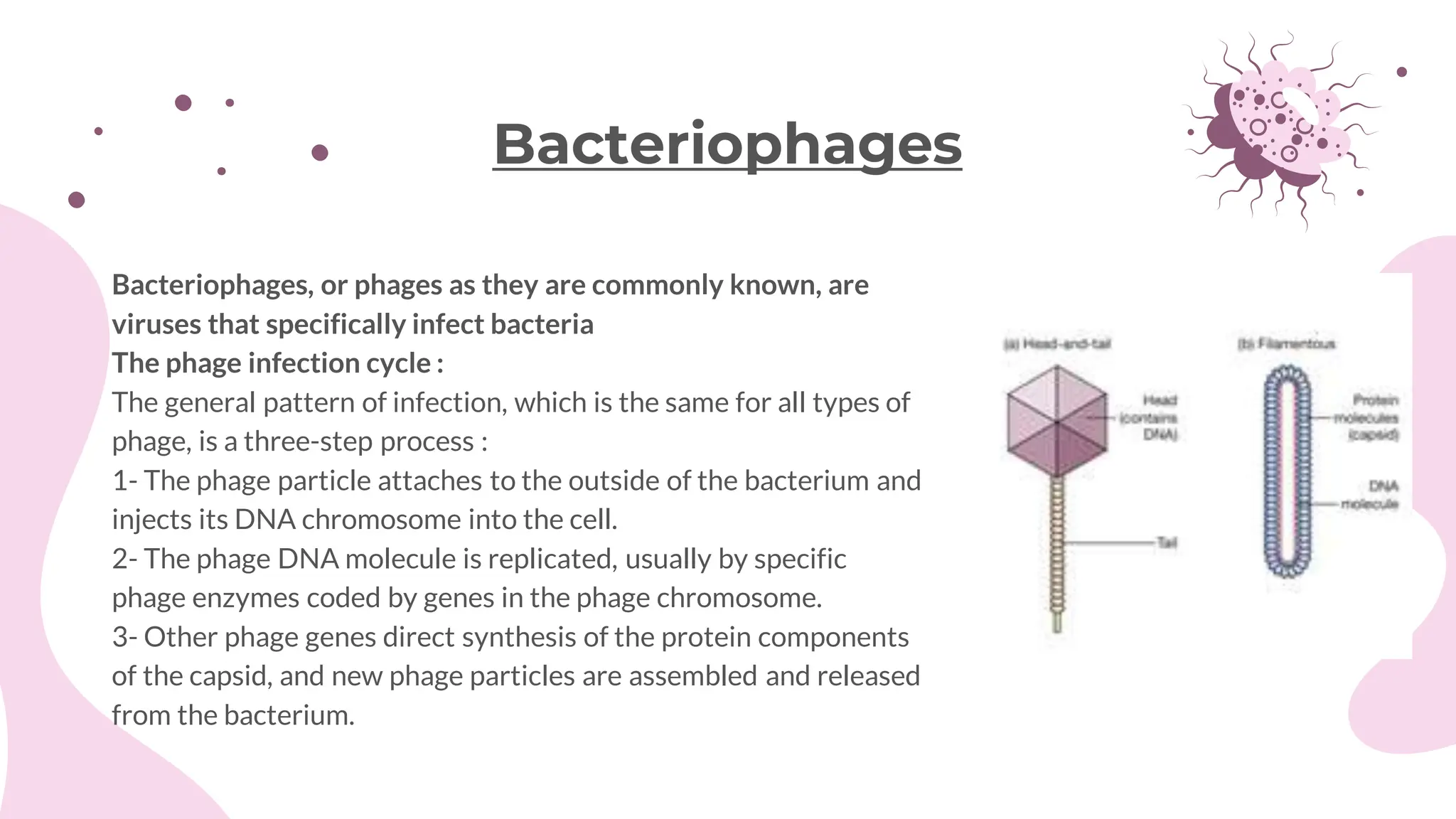
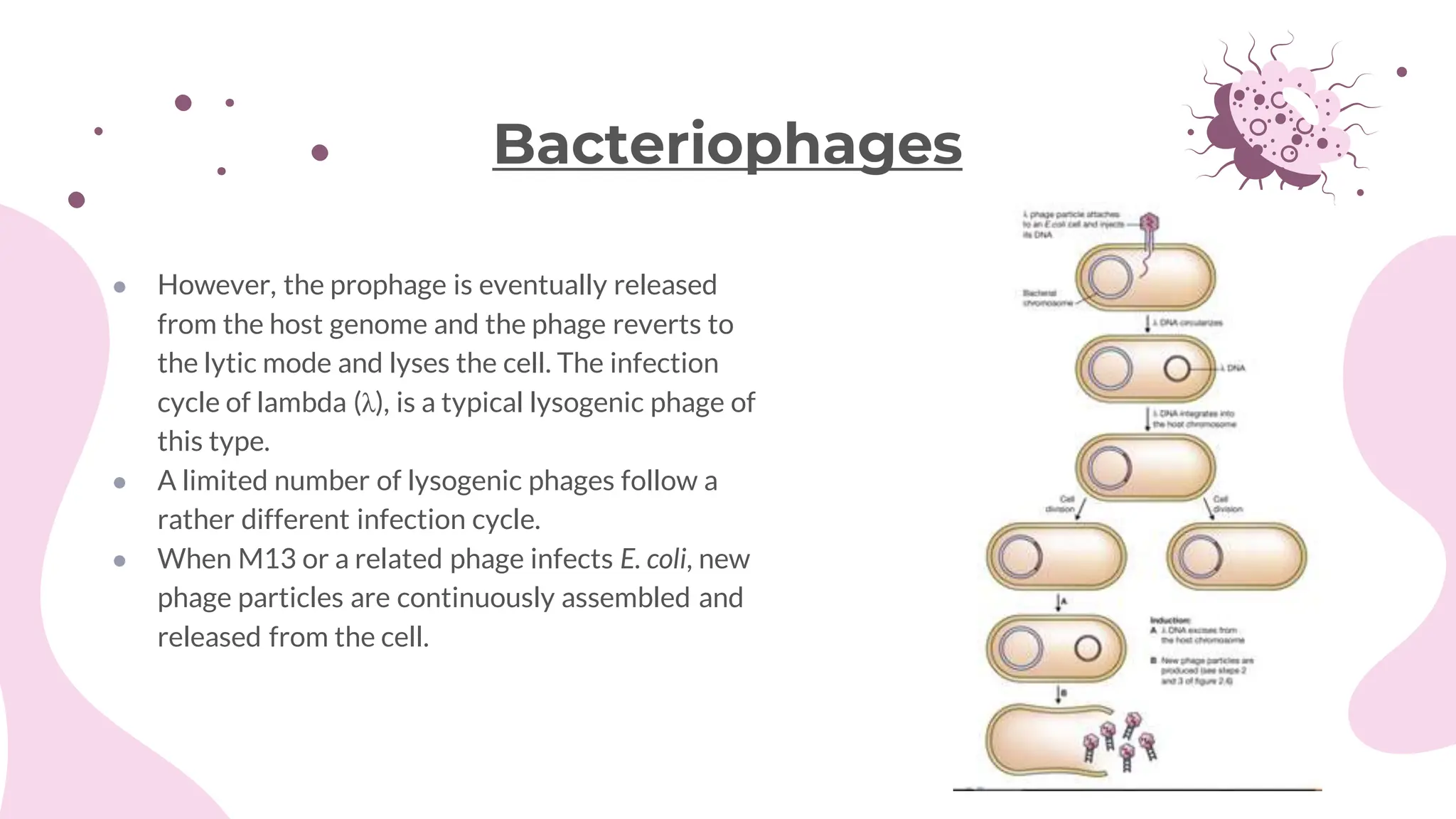
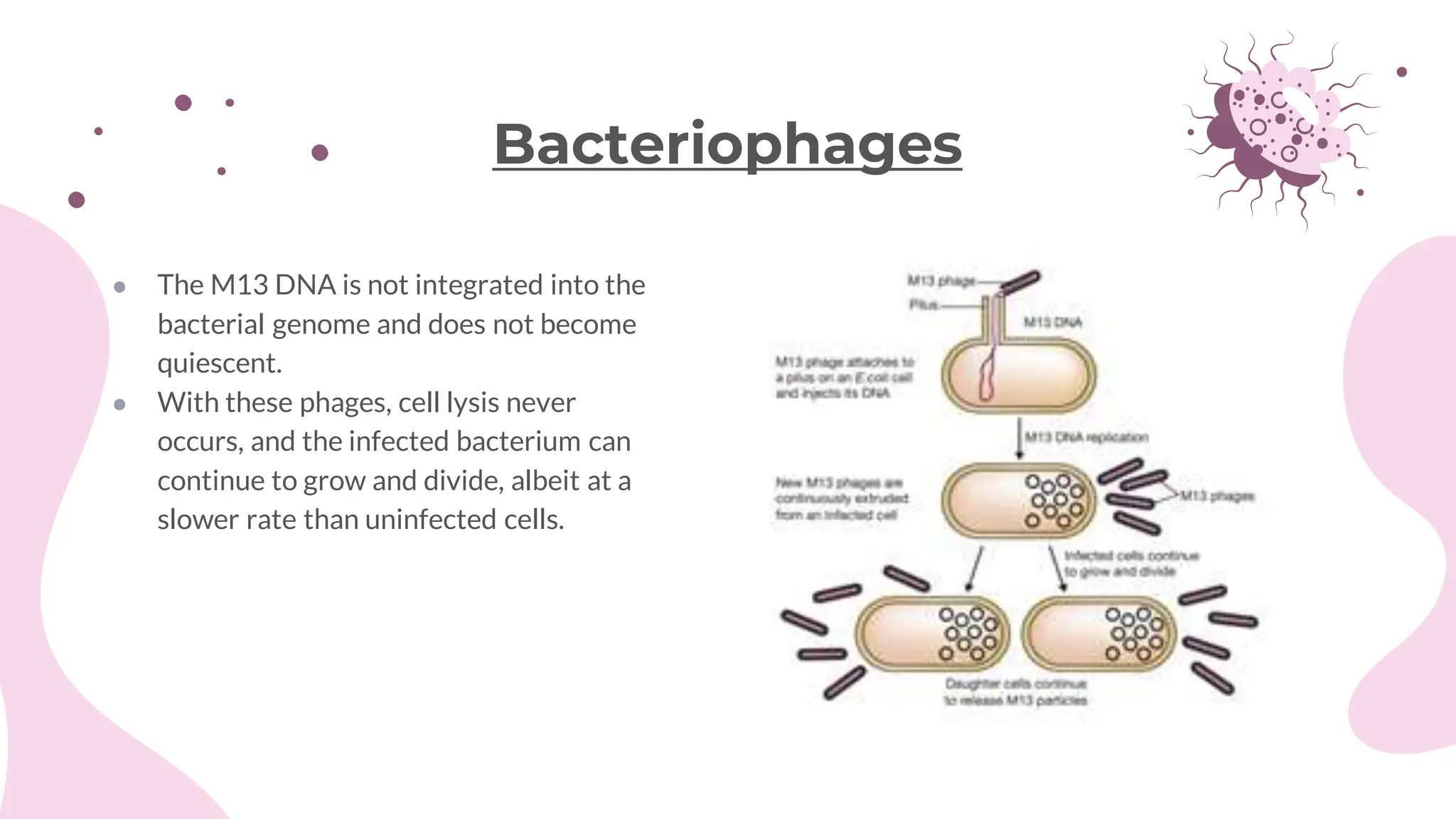
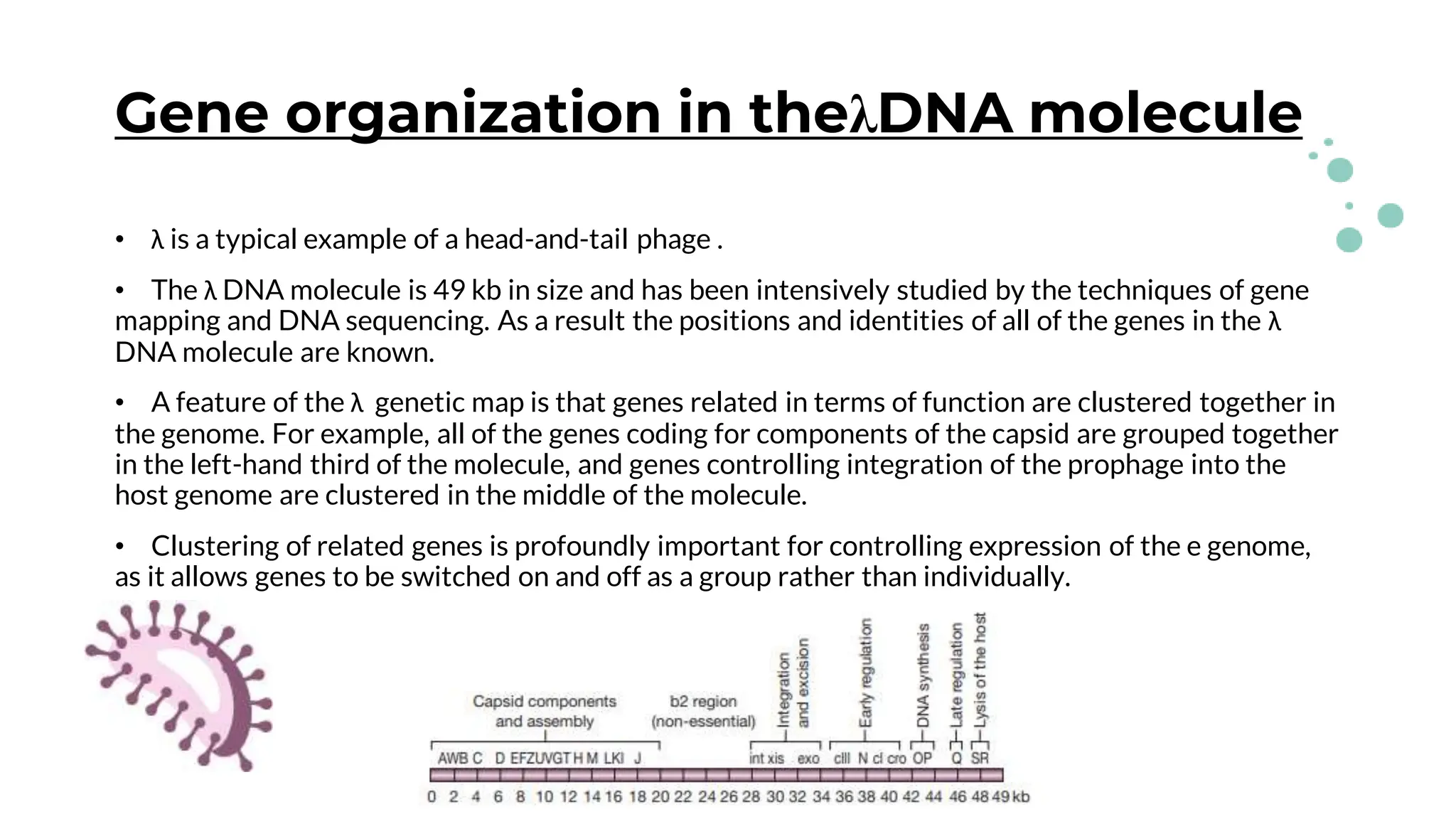
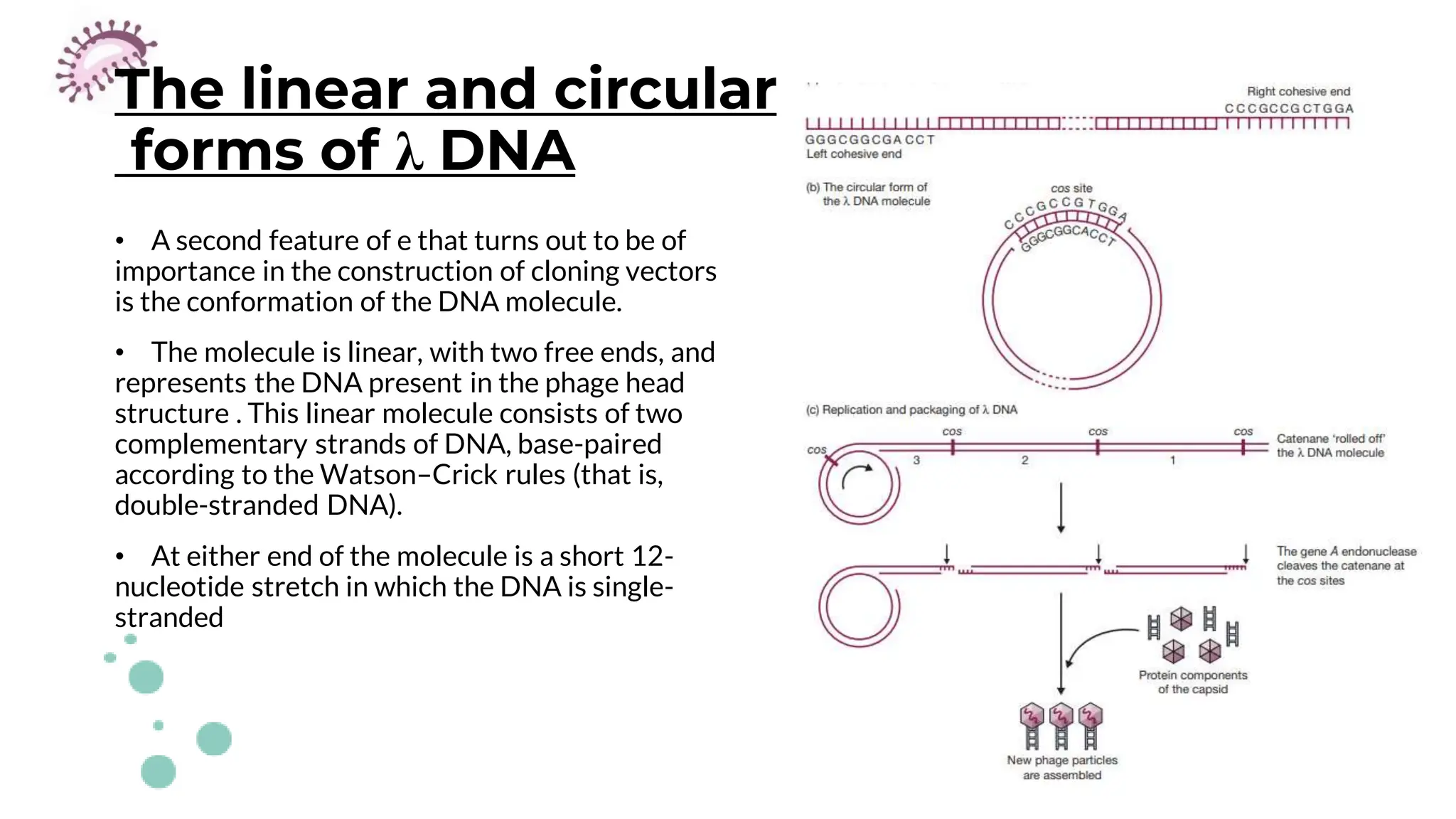
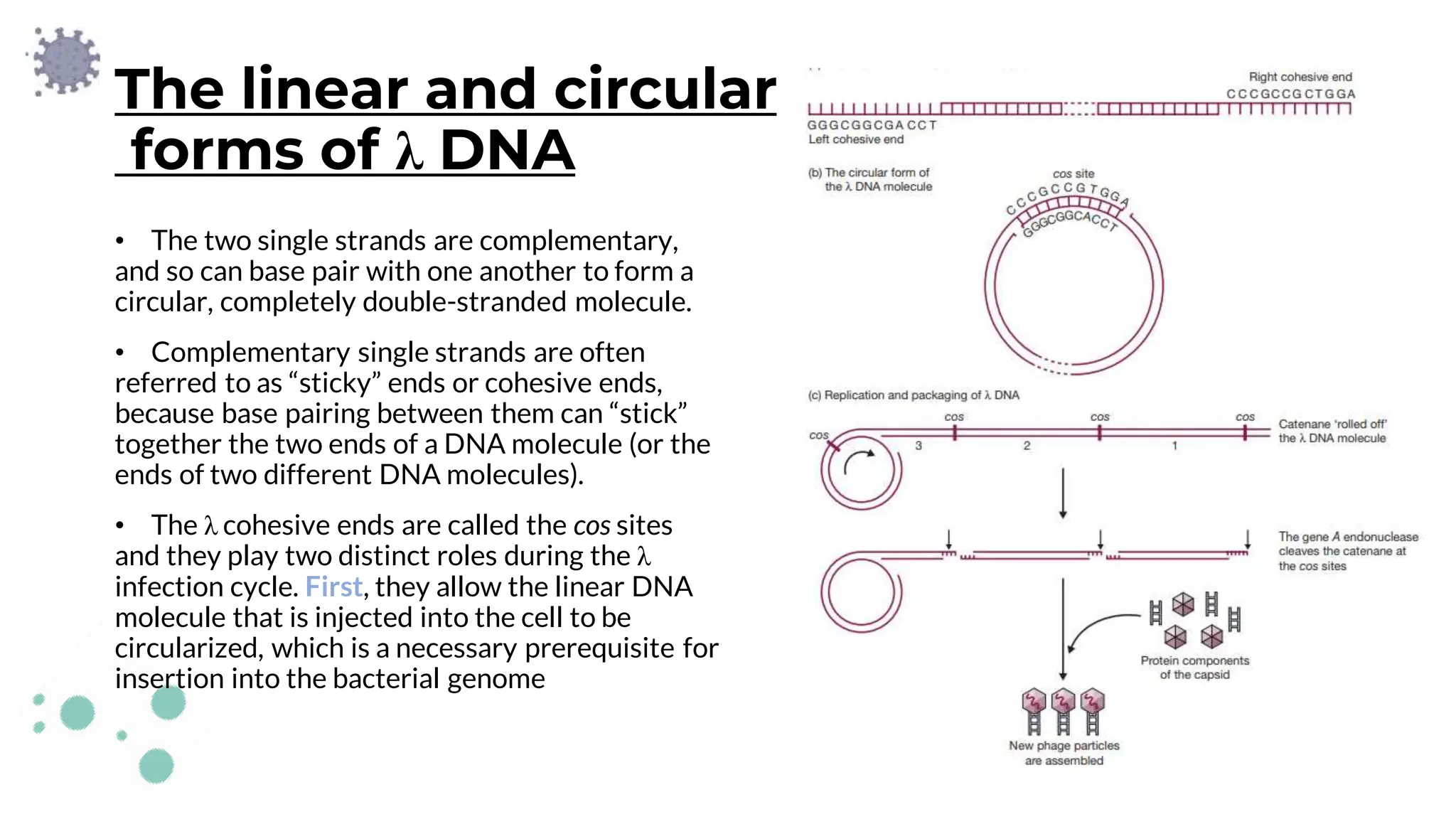
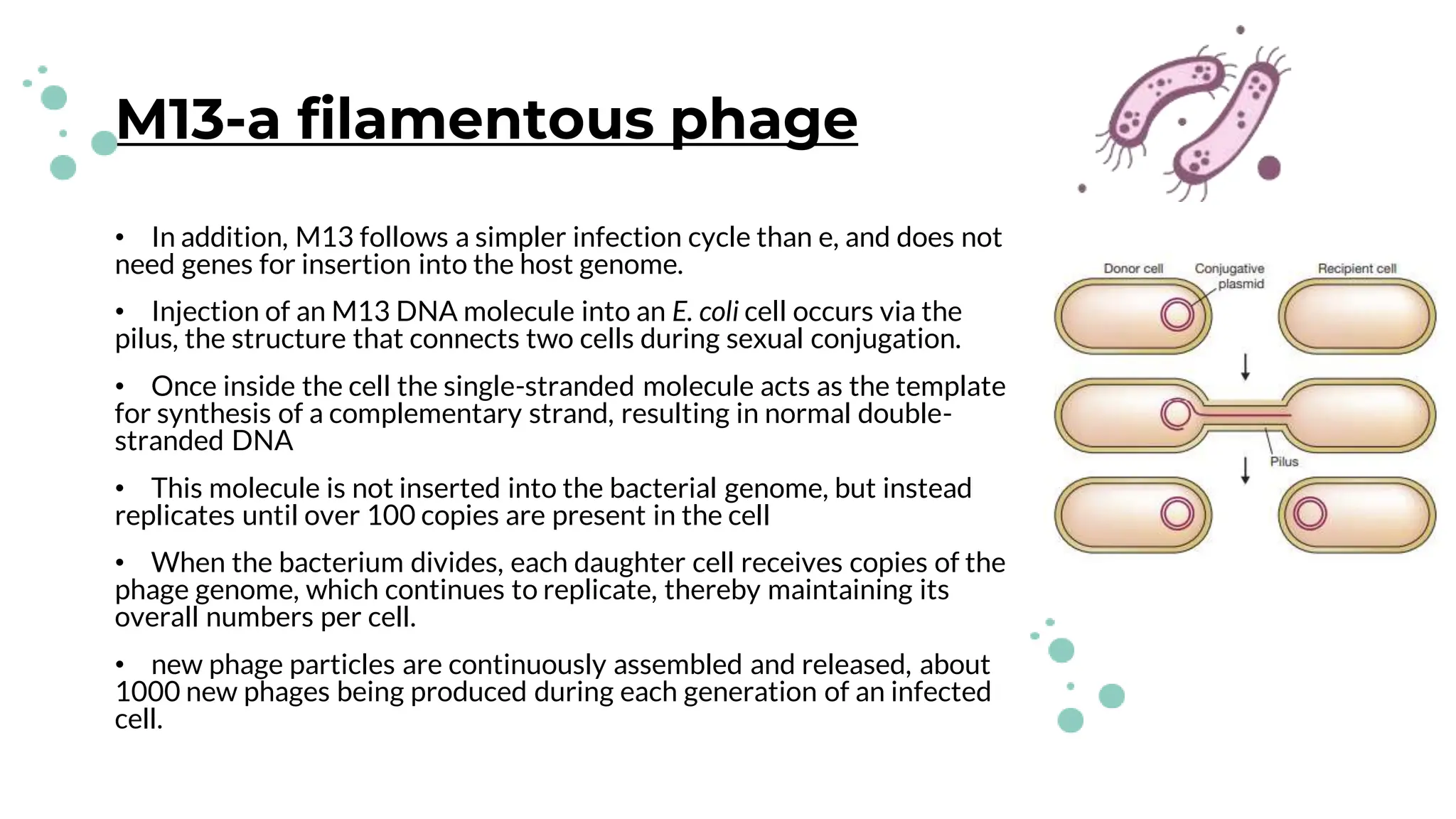
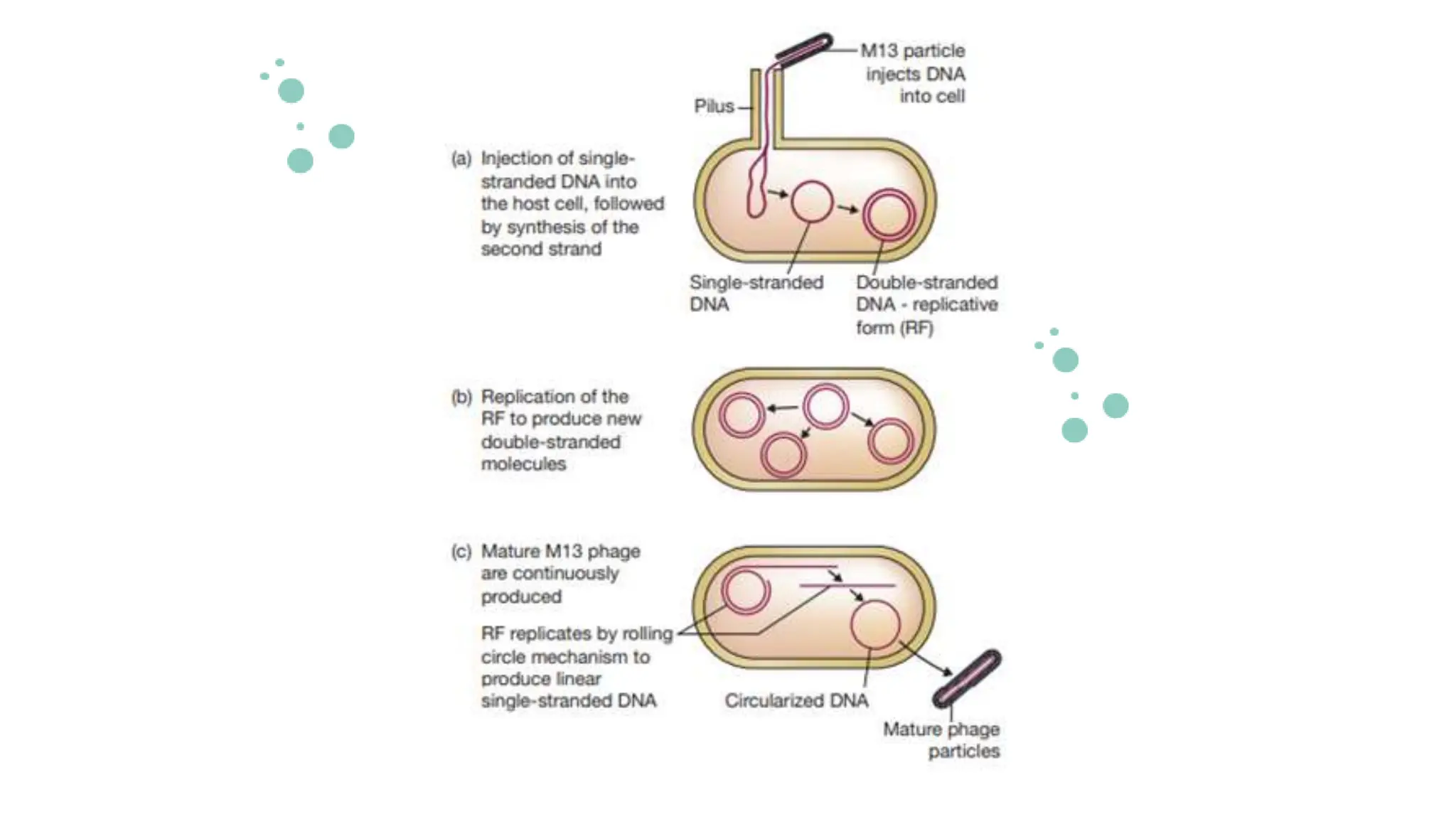
The document details the historical development and significance of gene cloning and DNA analysis, beginning with Gregor Mendel's work in inheritance and progressing through key discoveries in genetics, including the discovery of DNA as the genetic material. It explains the processes of gene cloning and polymerase chain reaction (PCR), highlighting their roles in isolating genes, facilitating DNA analysis, and their applications in medicine and forensics. The document also discusses vectors for gene cloning, particularly plasmids and bacteriophages, outlining their characteristics and importance in genetic engineering.