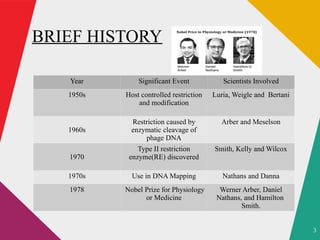
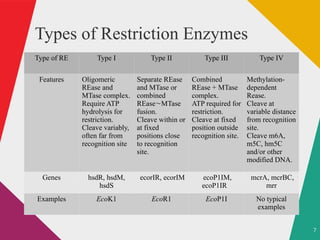
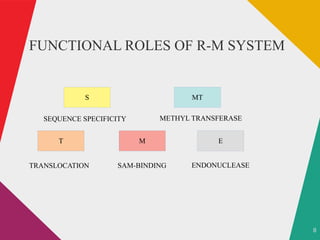
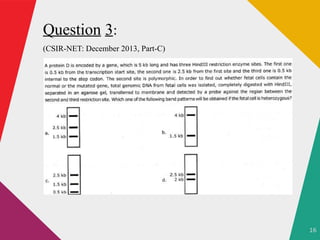
The document discusses restriction enzymes, which are molecular scissors that cut DNA at specific sites and are found in prokaryotes for defense. It details the history, types (I, II, III, IV), and functional roles of these enzymes, highlighting the significance of type II restriction enzymes in genetic manipulation and their applications. The document also includes nomenclature, mechanisms, and references for further reading.