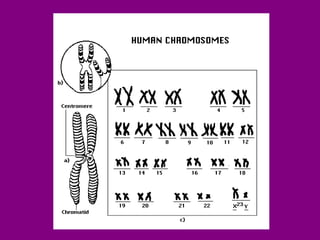
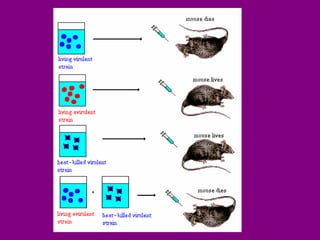
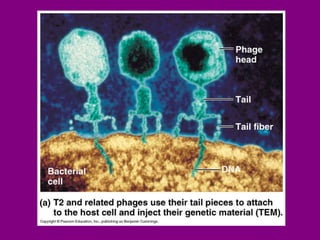
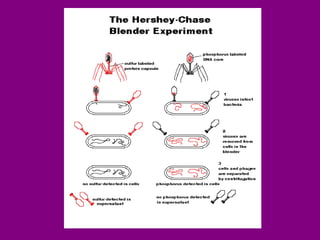
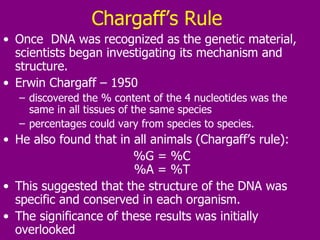
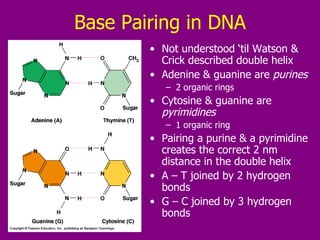
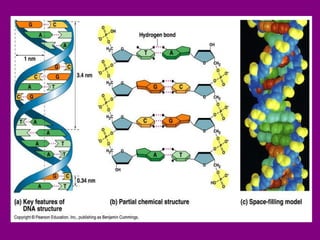
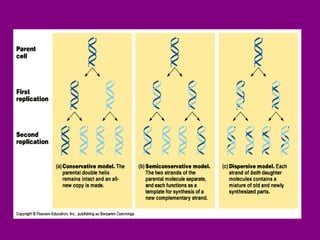
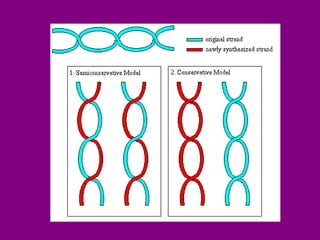
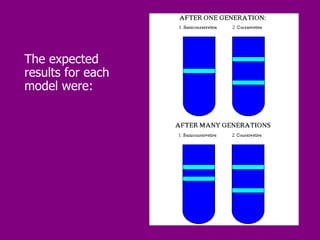
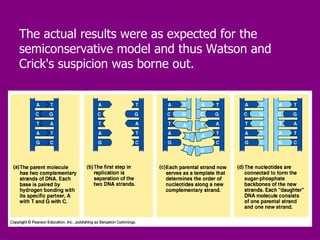
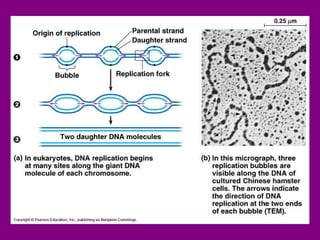
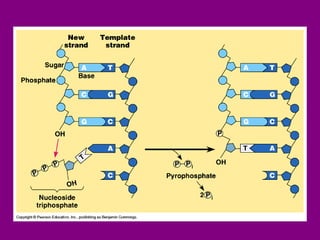
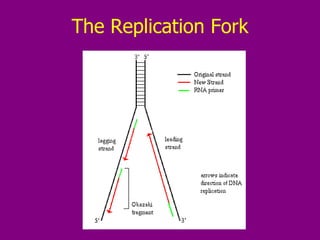
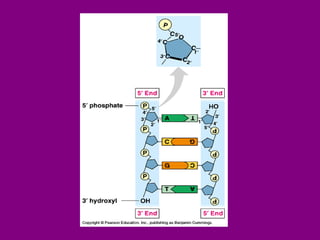
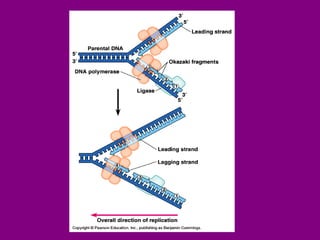
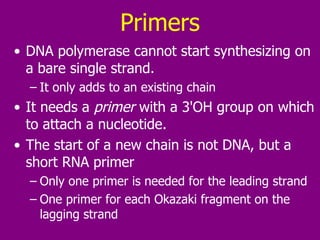
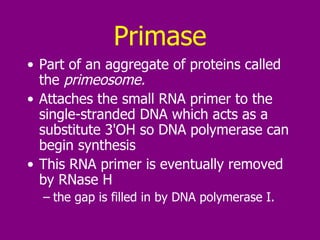
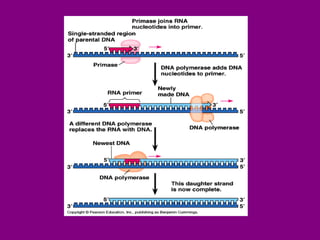
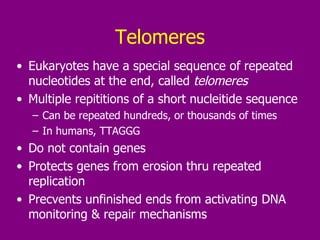
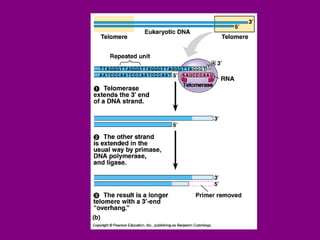
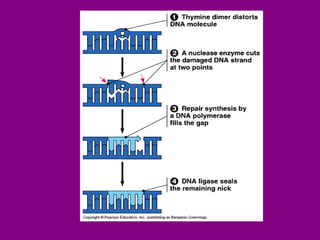
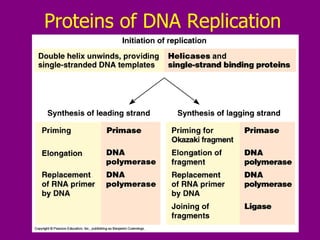
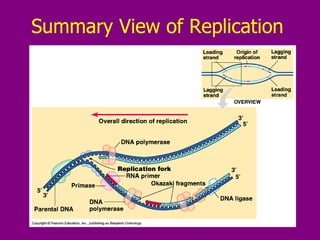
The document summarizes the central dogma of biology and the discovery of DNA as the genetic material. It describes key experiments that showed DNA replicates in a semiconservative manner, with each parental strand serving as a template for a new complementary daughter strand. The process of DNA replication requires several enzymes including DNA polymerase, helicase, ligase and primase to unwind, copy and join new DNA strands.