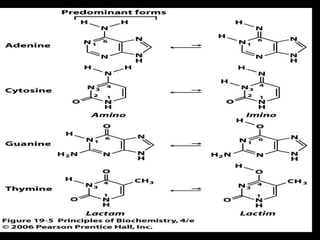
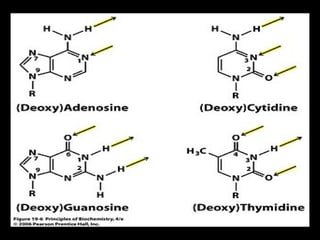
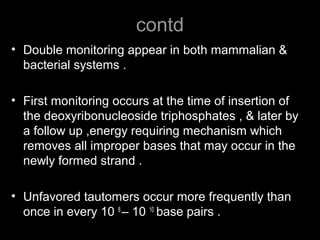
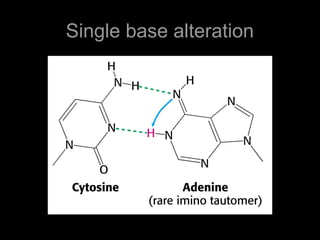
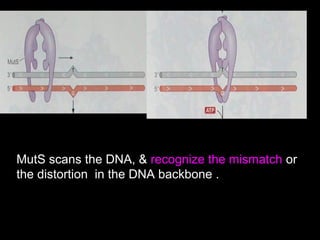
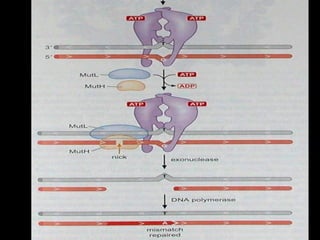
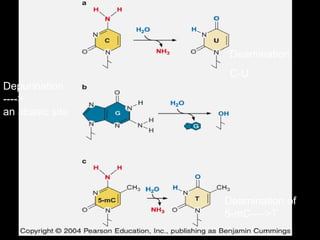
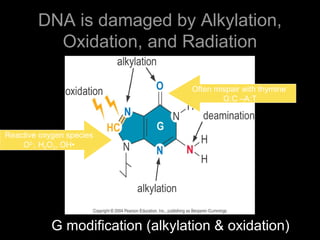
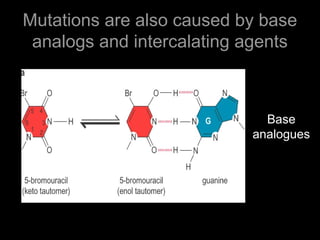
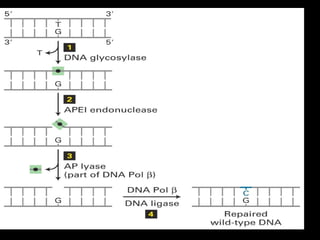
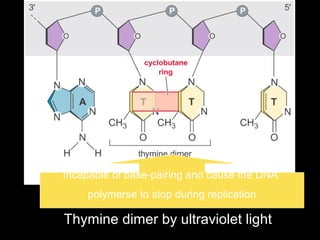
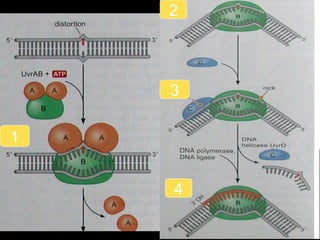
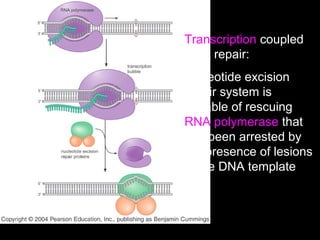
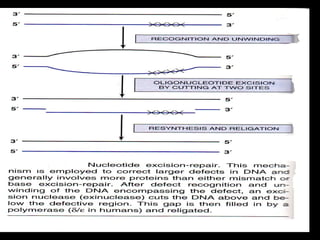
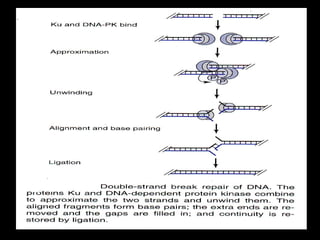
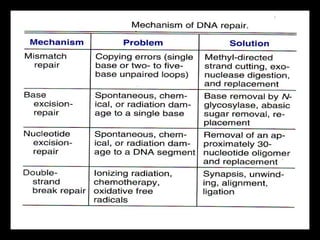
DNA repair mechanisms are essential for maintaining genomic integrity. There are several pathways for repairing different types of DNA damage: mismatch repair fixes errors during DNA replication, base excision repair removes damaged bases, nucleotide excision repair replaces larger sections of damaged DNA, and double-strand break repair fixes breaks in both DNA strands. Defects in DNA repair genes can lead to increased cancer risks and genetic disorders like xeroderma pigmentosum and Fanconi anemia. Overall, DNA repair helps prevent mutations from being passed to new cells.