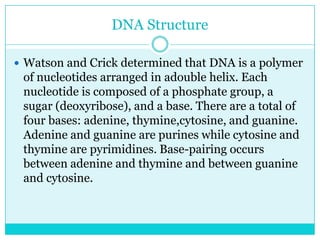
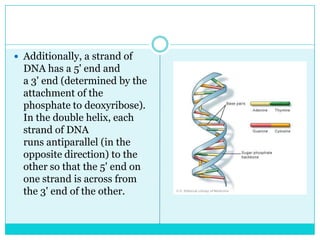
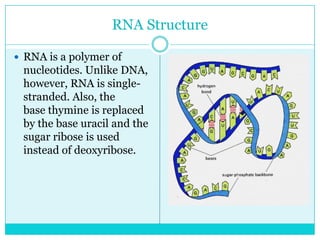
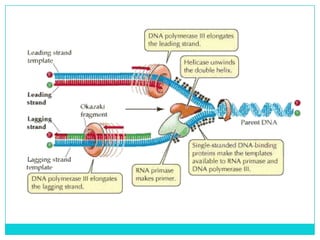
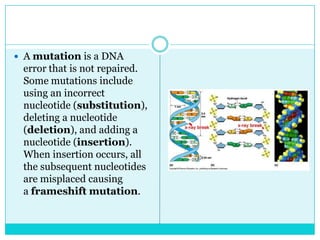
Watson and Crick determined DNA structure is a double helix of nucleotides with bases that pair together. DNA is replicated through a semi-conservative process where each old strand acts as a template for a new complementary strand. Errors during replication can lead to mutations in the DNA sequence. During protein synthesis, the DNA code is transcribed into mRNA which is then translated by ribosomes into a polypeptide chain according to the base sequence of codons.