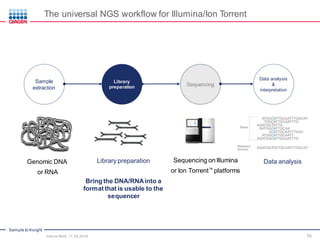
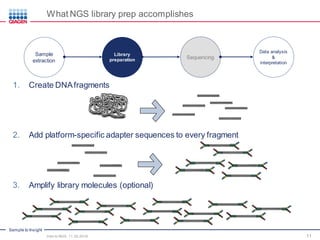
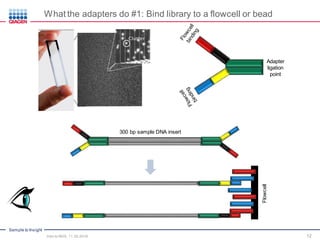
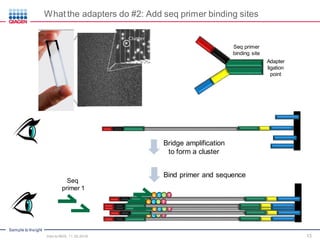
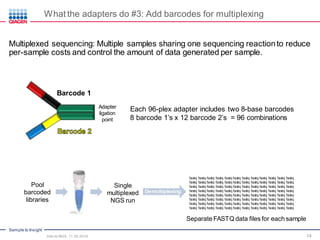
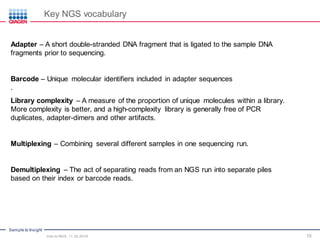
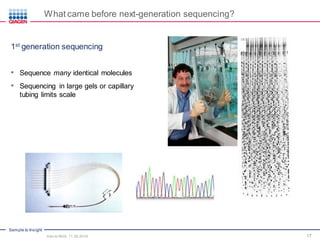
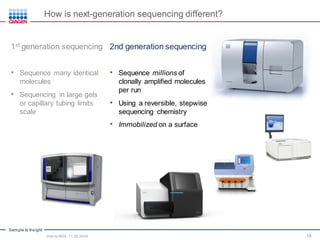
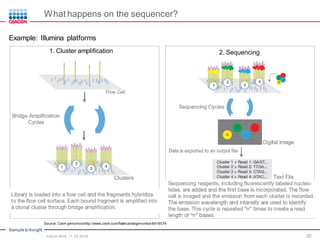
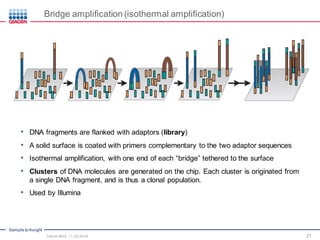
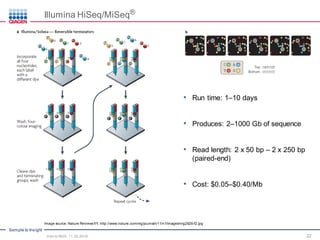
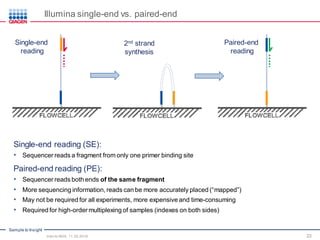
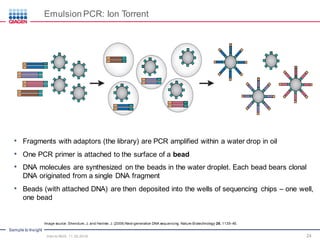
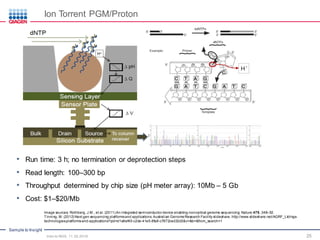
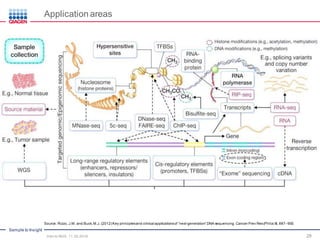
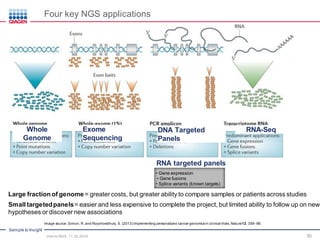
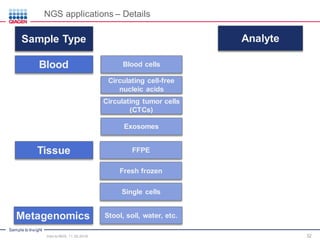
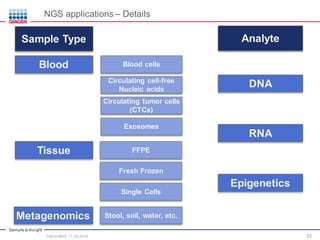
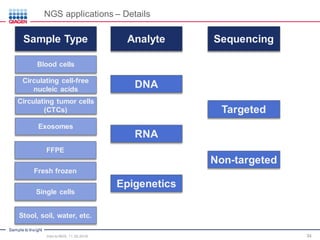
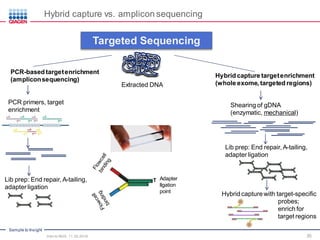
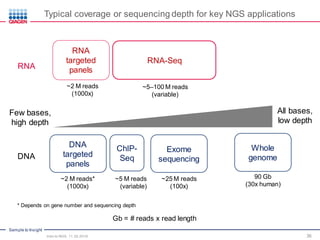
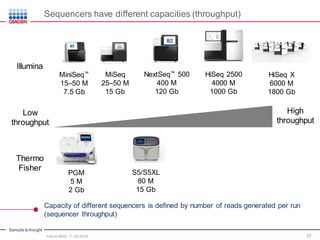
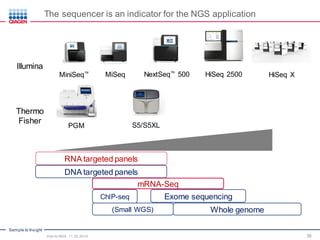
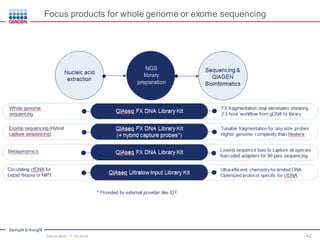
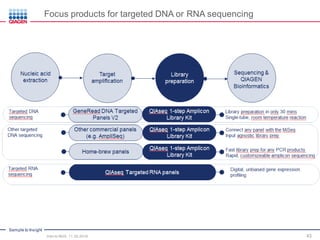
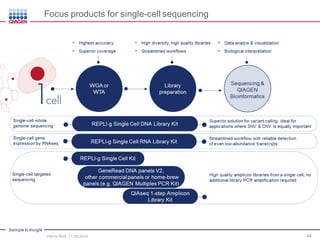
The document serves as an introduction to Next-Generation Sequencing (NGS), providing an overview of NGS technologies, library preparation methods, and applications. It discusses the evolution from first-generation to third-generation sequencing, highlights the NGS workflow, and covers various applications including whole genome sequencing, exome sequencing, and targeted panels. Additionally, it presents the Qiagen portfolio of NGS products and resources for further learning.