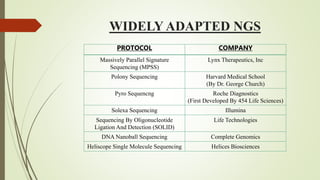
Next-generation sequencing (NGS), also known as high-throughput sequencing, is a modern and cost-effective technology for sequencing nucleic acids, significantly improving upon traditional Sanger sequencing. It enables the simultaneous sequencing of millions of DNA or RNA fragments through methodologies including template preparation, sequencing, imaging, and data analysis. NGS has applications across various fields such as genomics, virology, epidemiology, and cancer studies.