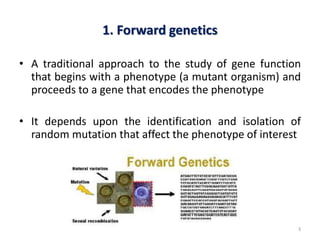
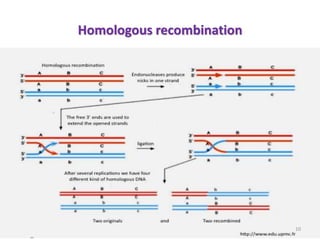
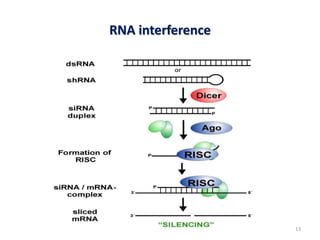
This document discusses forward and reverse genetic approaches for understanding gene function. Forward genetics begins with a phenotype and identifies the underlying gene, while reverse genetics starts with a gene and determines its phenotype. Specific reverse genetic techniques described include large-scale random mutagenesis, homologous recombination, transposable element excision, RNA interference, genome editing using ZFNs/TALENs/CRISPR, and site-directed mutagenesis combined with transgenics. The document provides details on how each technique is used to alter genes and study their function.