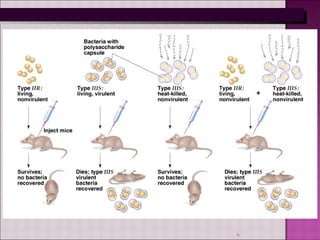
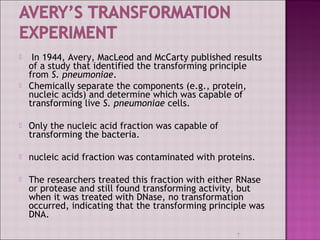
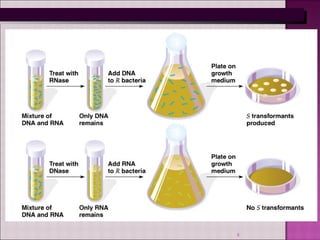
1. Experiments in the 1940s-1950s provided evidence that DNA, not protein, is the genetic material. This included Avery, MacLeod and McCarty's experiment showing only DNA could transform bacteria.
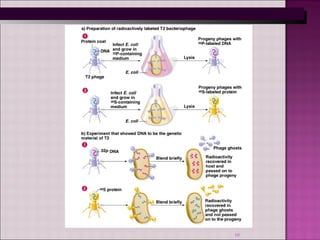
2. Further evidence came from Hershey and Chase's experiment labelling bacterial virus DNA and protein differently, finding that DNA entered host bacteria cells while protein did not.
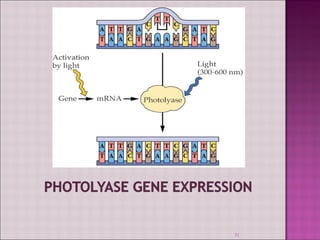
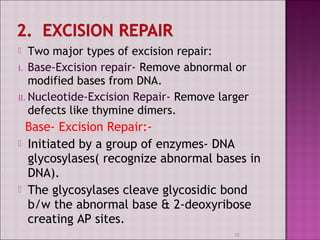
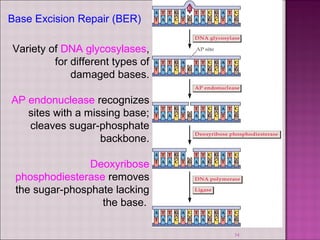
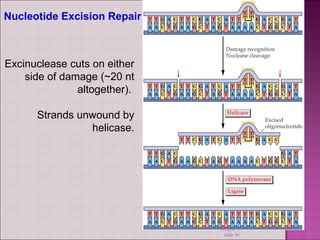
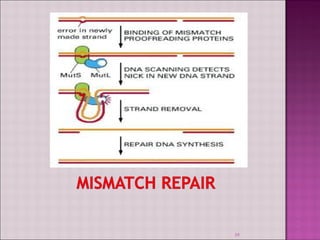
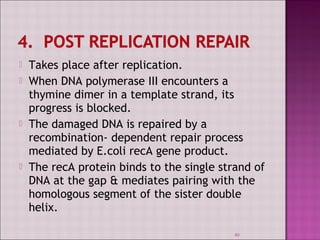
3. DNA can be damaged by various agents like radiation, chemicals, and oxidation. Cells have multiple DNA repair mechanisms like base-excision repair and nucleotide-excision repair to repair damages and prevent mutations.