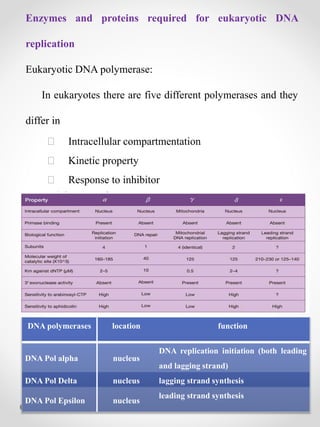
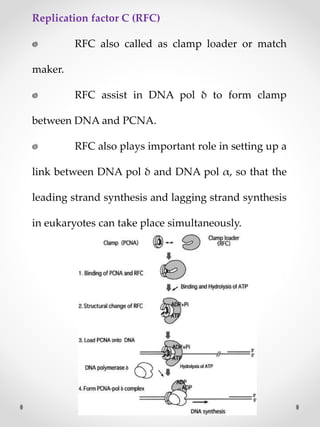
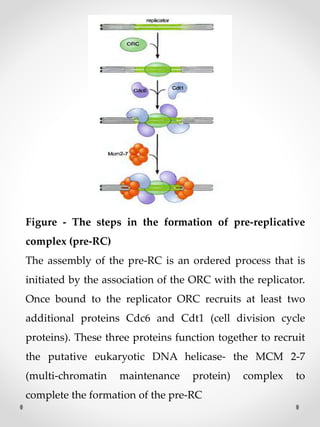
The document summarizes eukaryotic DNA replication. It discusses that DNA replication in eukaryotes is more complex than prokaryotes due to larger genome size and chromatin packaging. The key stages of eukaryotic replication are similar to prokaryotes, including origin of replication, formation of replication forks, semiconservative replication and synthesis of leading and lagging strands. However, eukaryotic replication involves additional proteins and is slower due to chromatin remodeling required to access DNA.