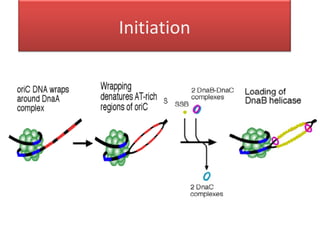
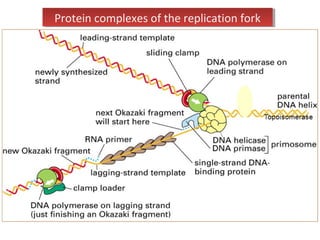
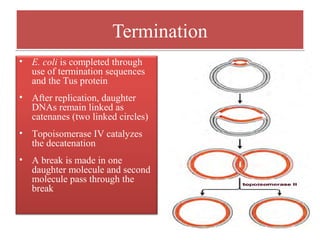
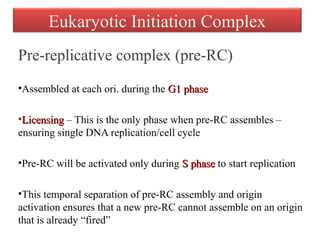
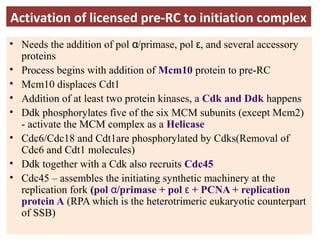
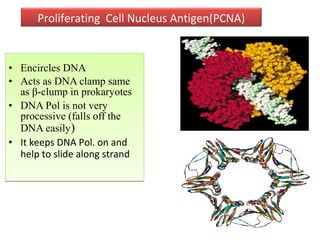
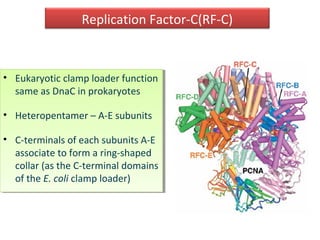
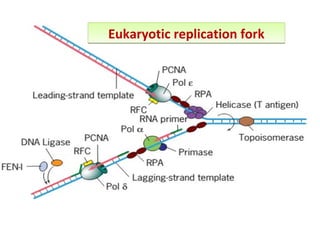
DNA replication is the process by which DNA copies itself for transmission to daughter cells. In the late 1950s, three models were proposed for how DNA replicates: conservative, semi-conservative, and dispersive. Experiments showed that the semi-conservative model is correct, where each parental DNA strand acts as a template to replicate a new partner strand. DNA replication requires DNA and RNA polymerases, helicase, topoisomerases, primase, ligase and other proteins. It occurs through initiation, elongation and termination steps in both prokaryotes and eukaryotes, though eukaryotes have multiple replication origins and use RNA primers on the lagging strand.