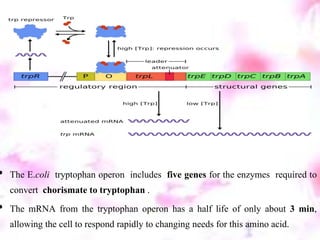
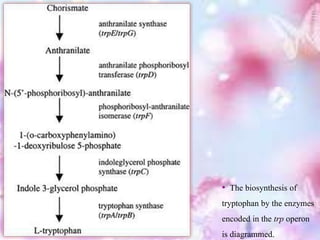
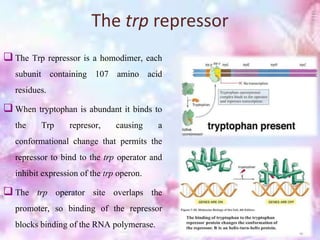
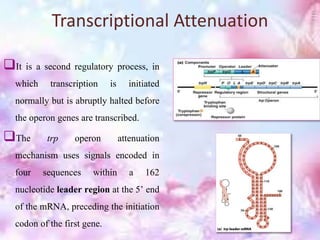
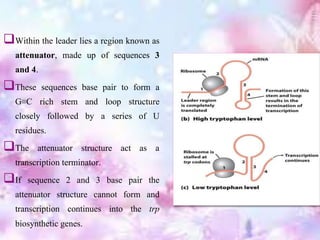
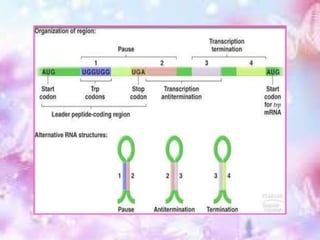
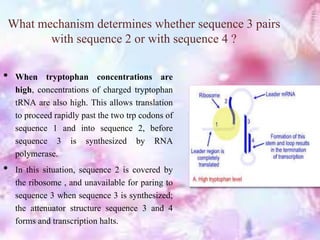
The tryptophan operon regulates the biosynthesis of tryptophan in E. coli through transcriptional attenuation and repression. It contains five genes encoding the enzymes needed to synthesize tryptophan. When tryptophan levels are high, the tryptophan repressor binds to the operator site, preventing transcription. Additionally, a regulatory region can form a terminator stem-loop structure to halt transcription if tryptophan tRNA levels are high during translation of the leader mRNA sequence. However, if tryptophan levels are low, the terminator structure does not form and transcription of the operon proceeds.