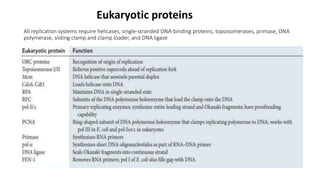
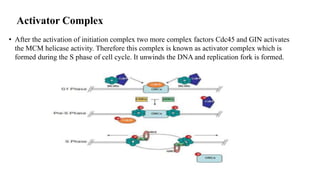
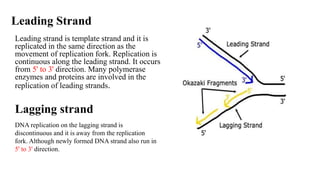
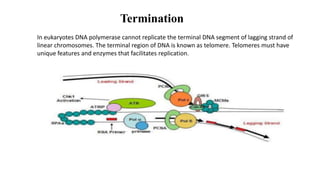
The document outlines the process of DNA replication in eukaryotes, highlighting differences from prokaryotic replication such as multiple origins of replication and slower rates of nucleotide incorporation. It details the roles of various DNA polymerases, the formation of pre-replication and initiation complexes, and the mechanics of elongation and termination. Key proteins and complexes involved in maintaining the structure and function of DNA during replication are also discussed.