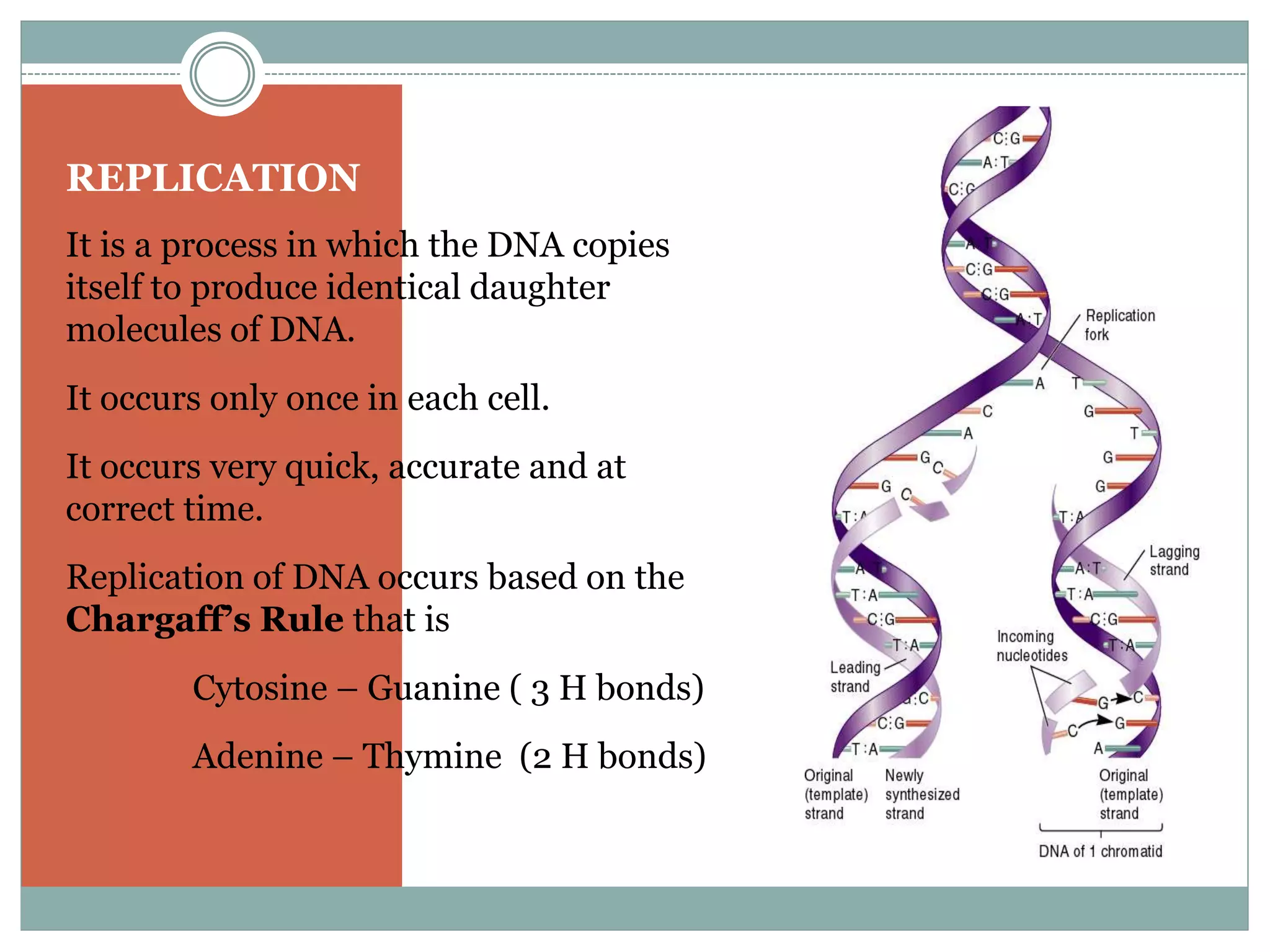
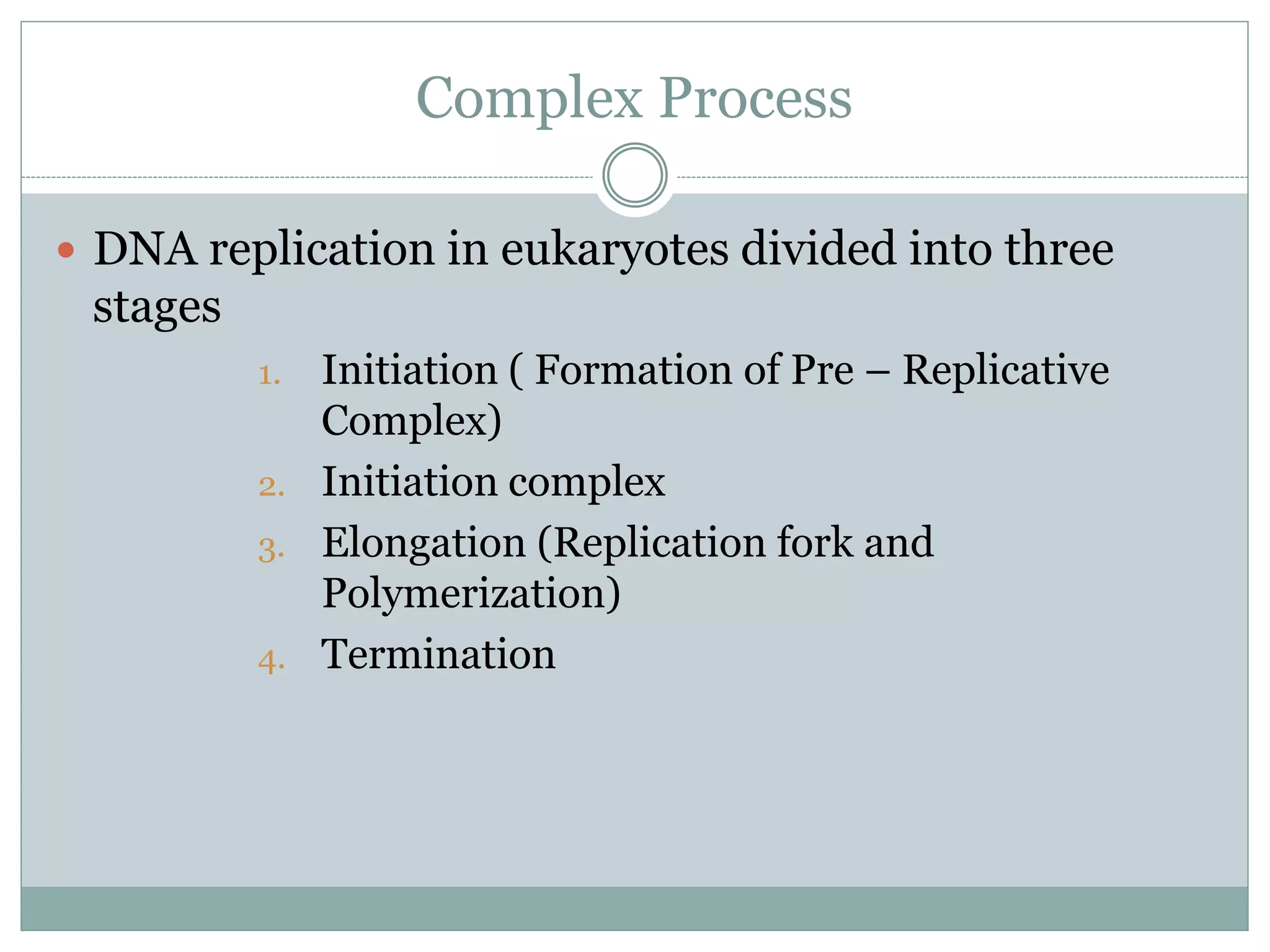
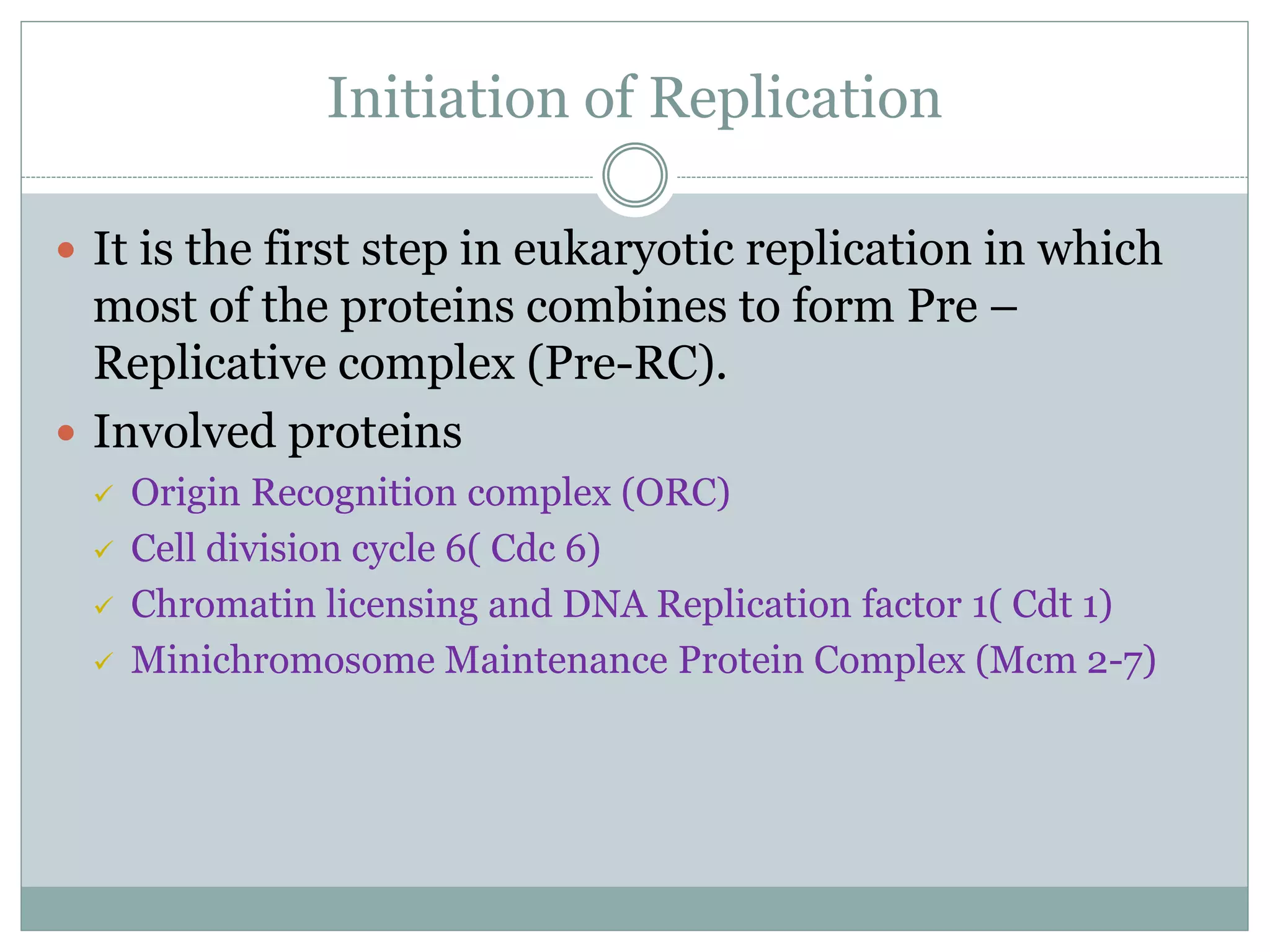
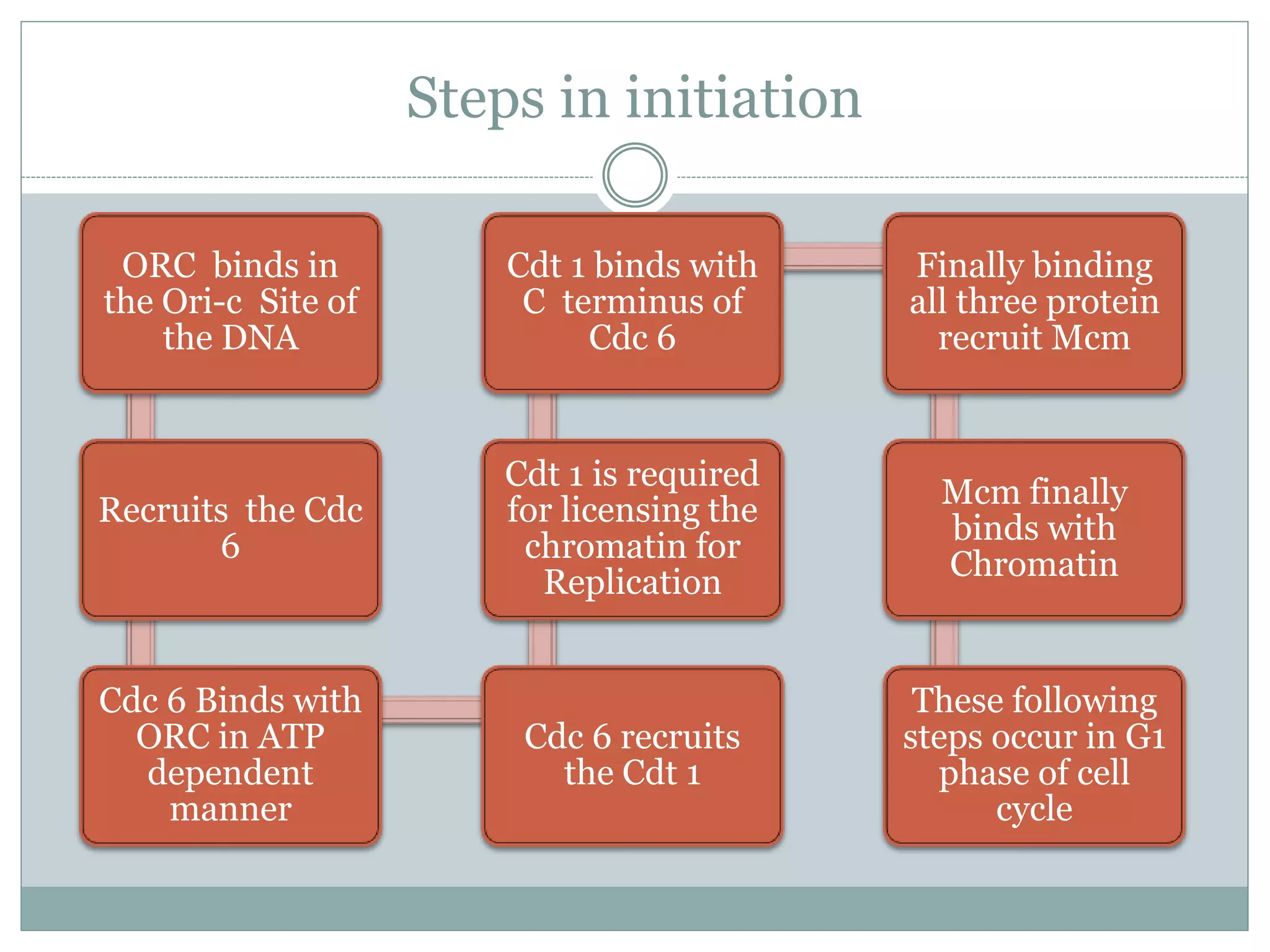
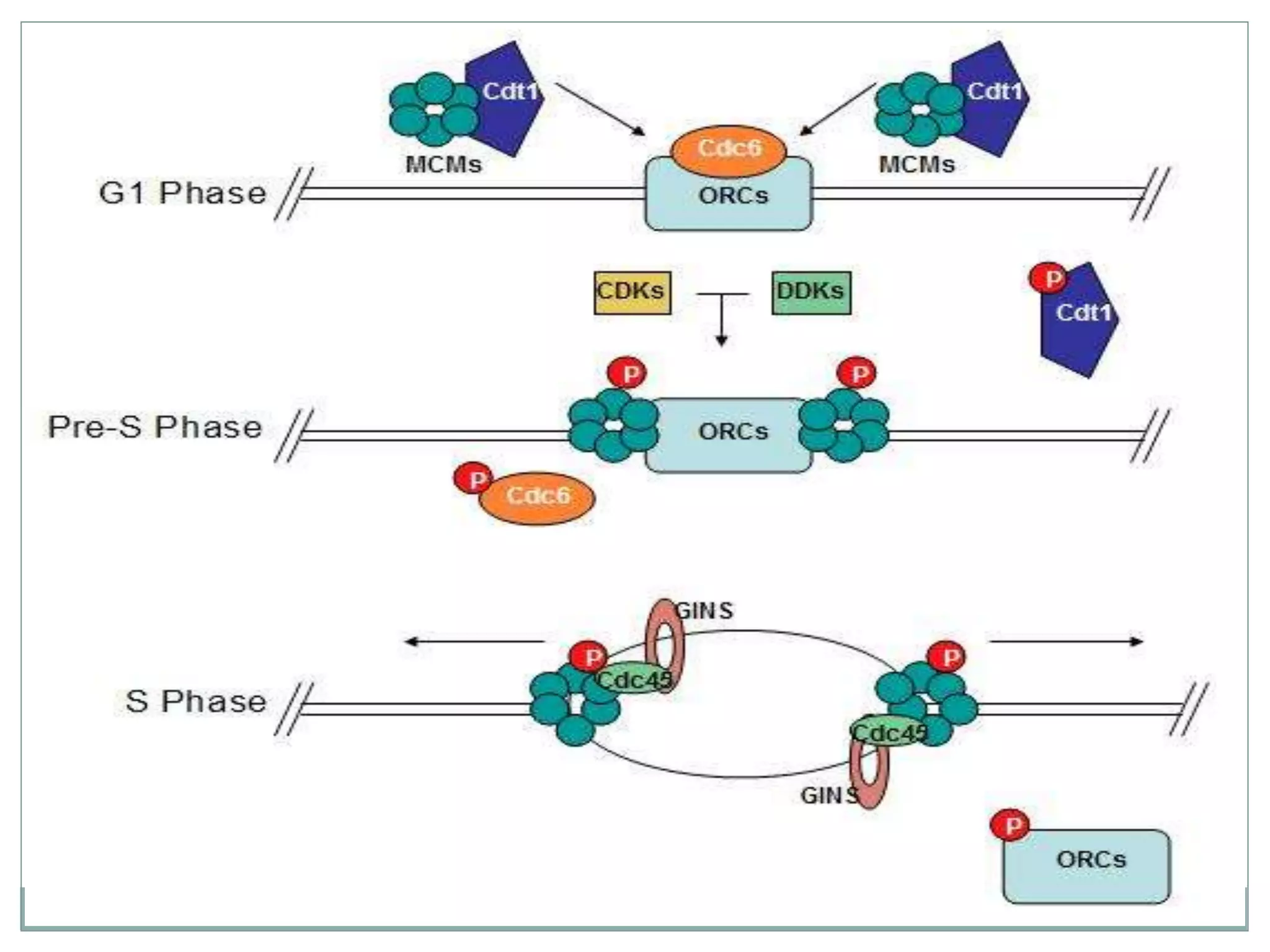
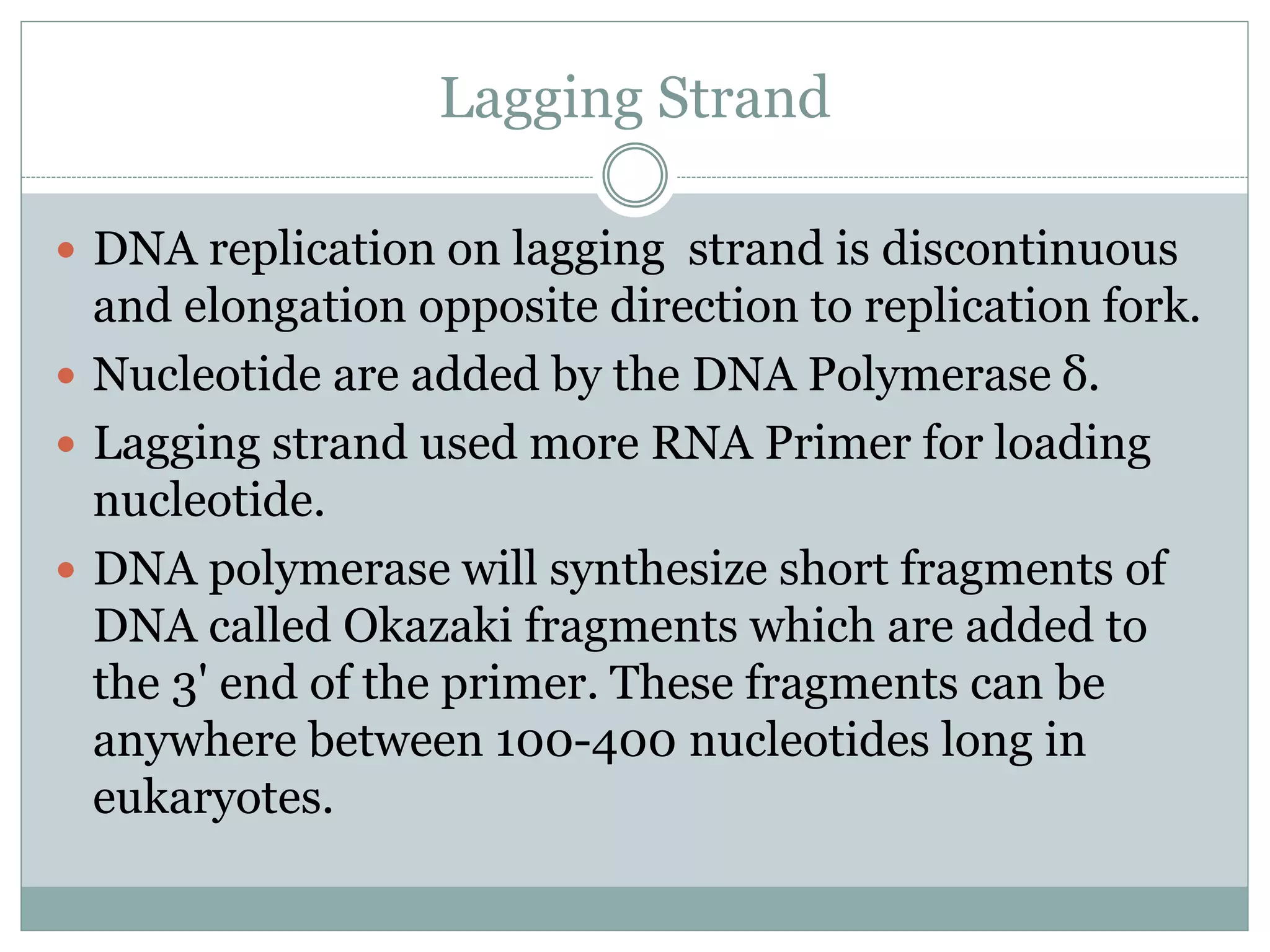
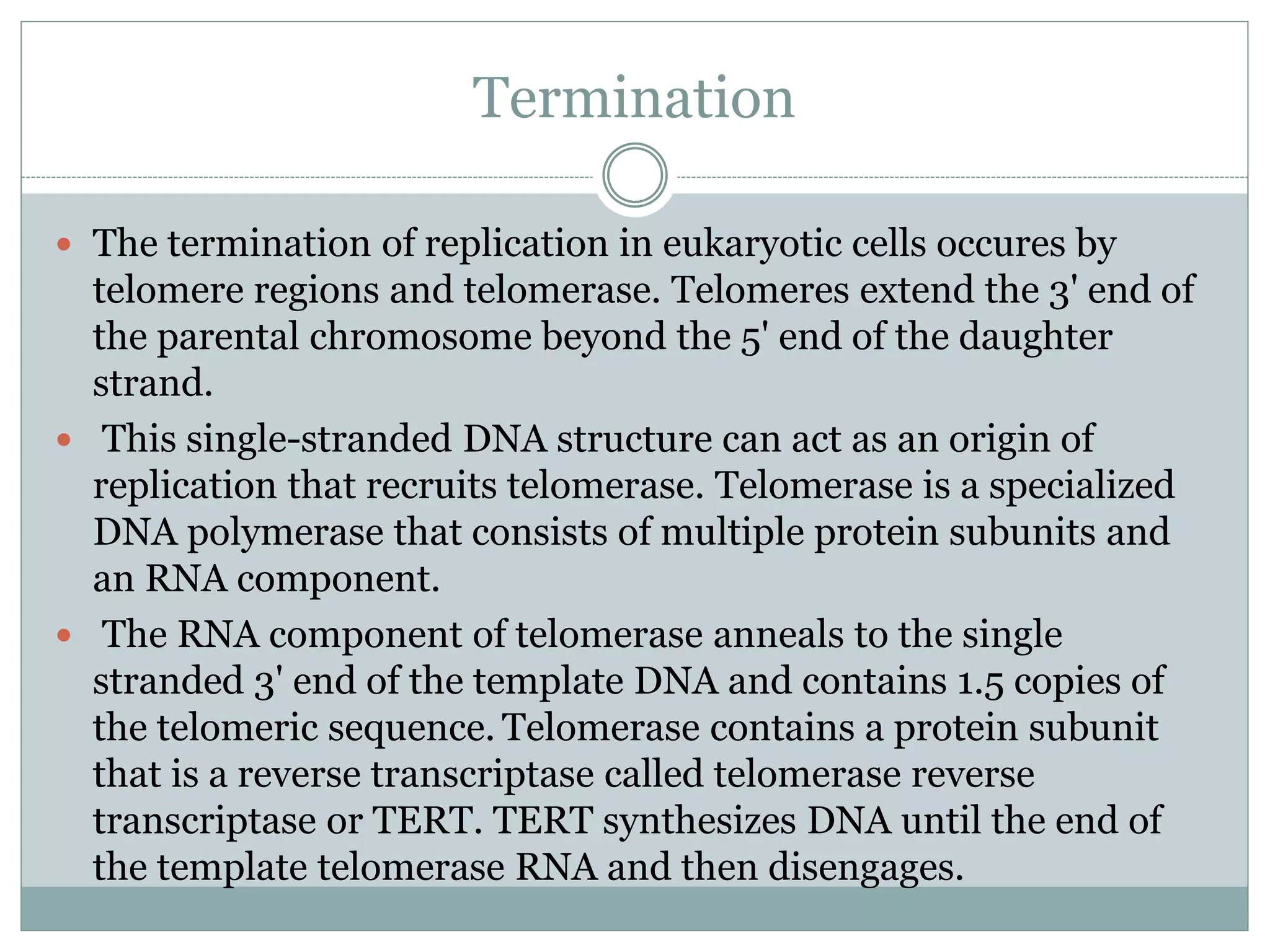
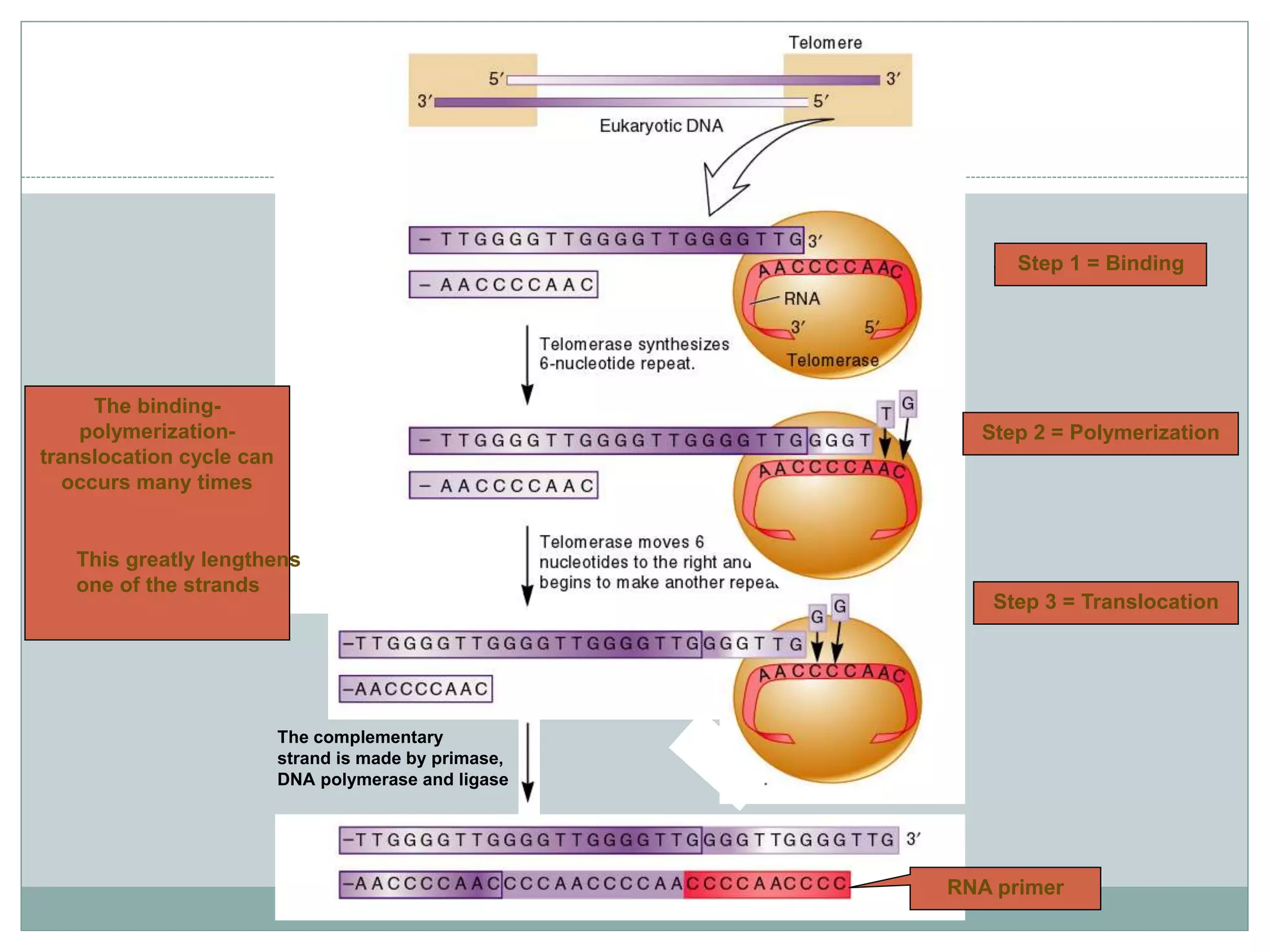
This document describes the process of DNA replication in eukaryotes. It occurs in S phase of the cell cycle and involves three main stages: initiation, formation of the initiation complex, and elongation. Initiation requires the assembly of pre-replication complexes containing ORC, Cdc6, Cdt1 and MCM proteins. In S phase, Cdc45 and GINS are recruited to form the initiation complex. Elongation proceeds bidirectionally from replication forks, with leading strand synthesis continuous and lagging strand discontinuous via Okazaki fragments. Replication terminates at telomeres.