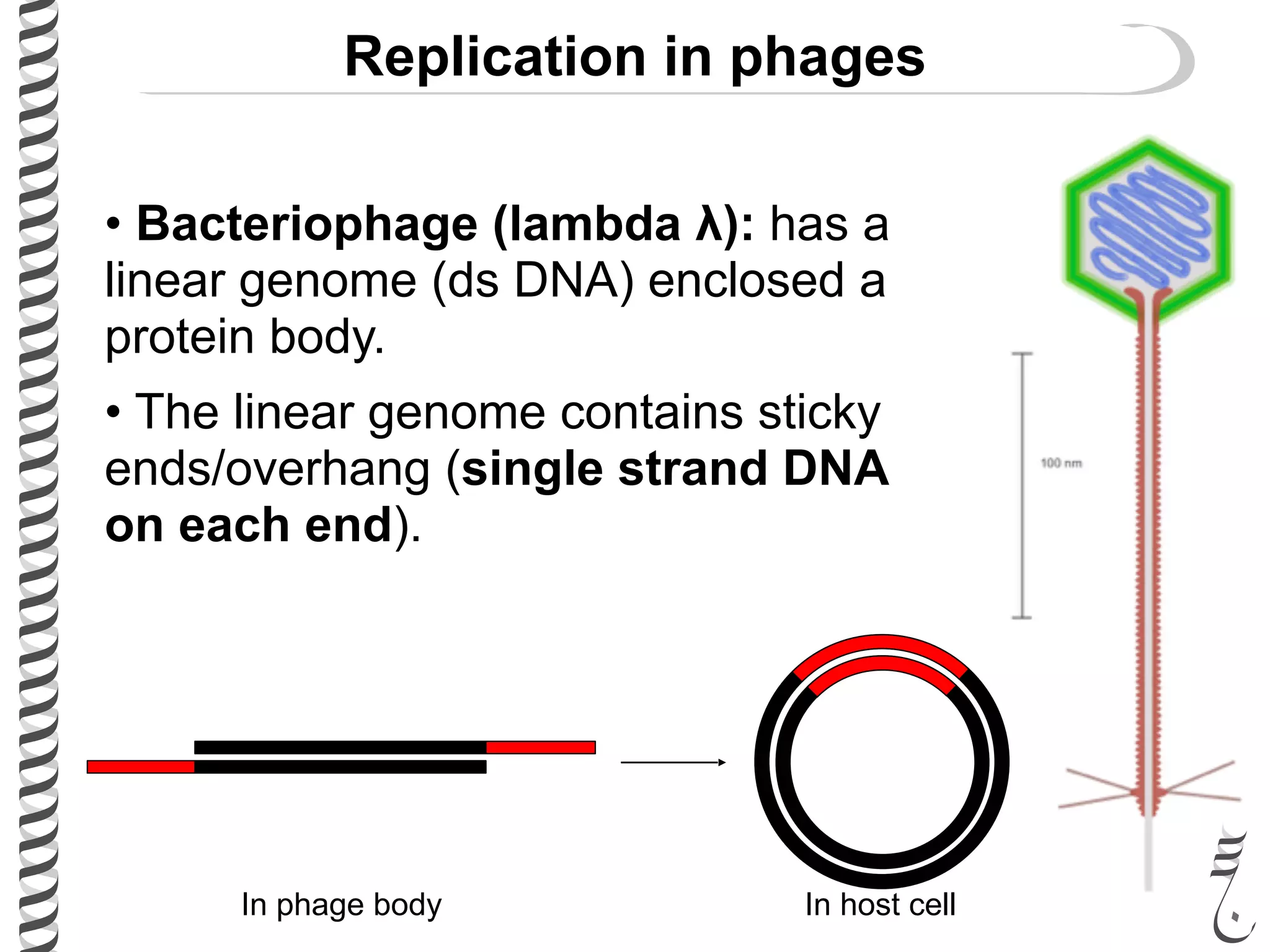
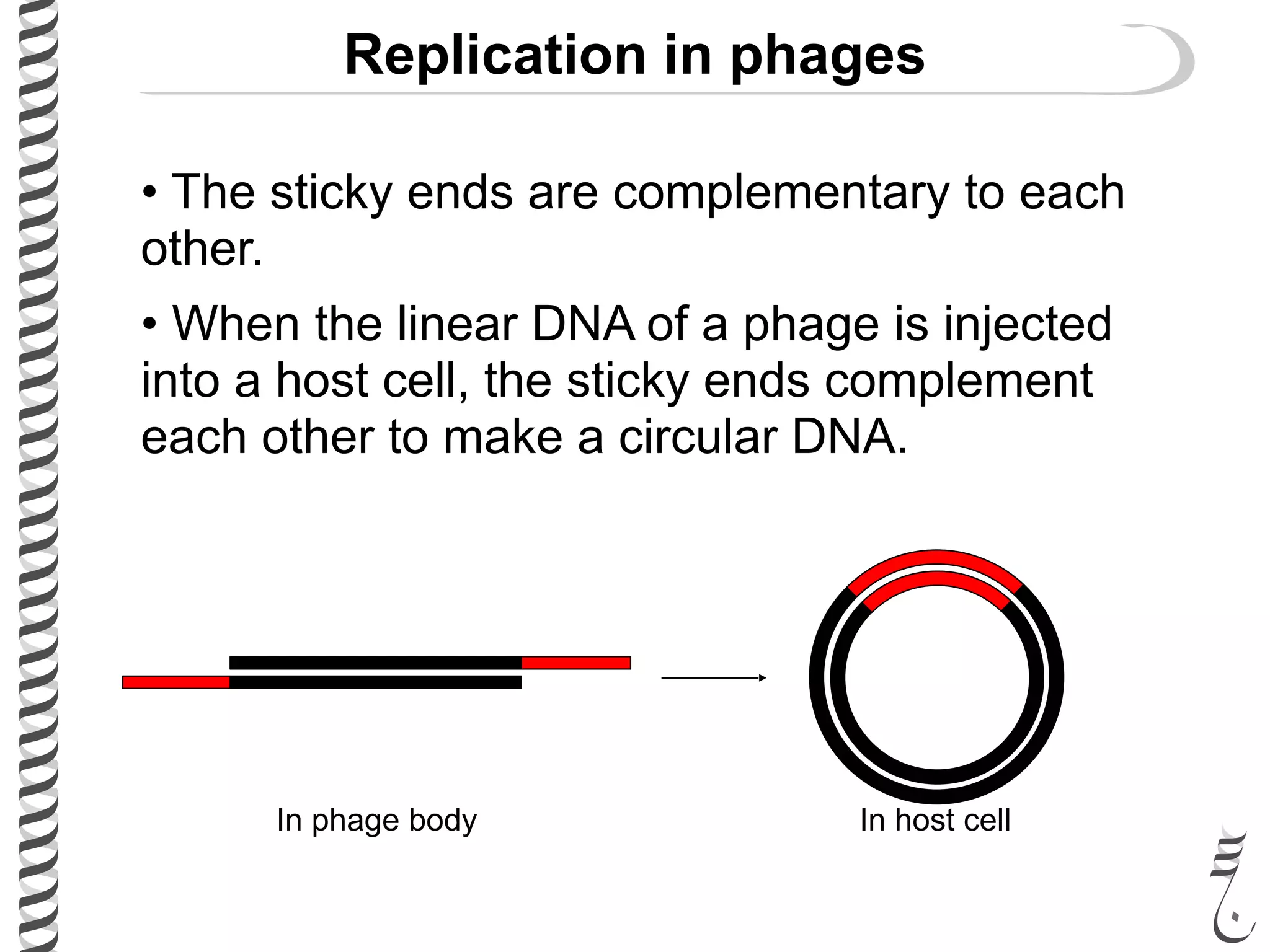
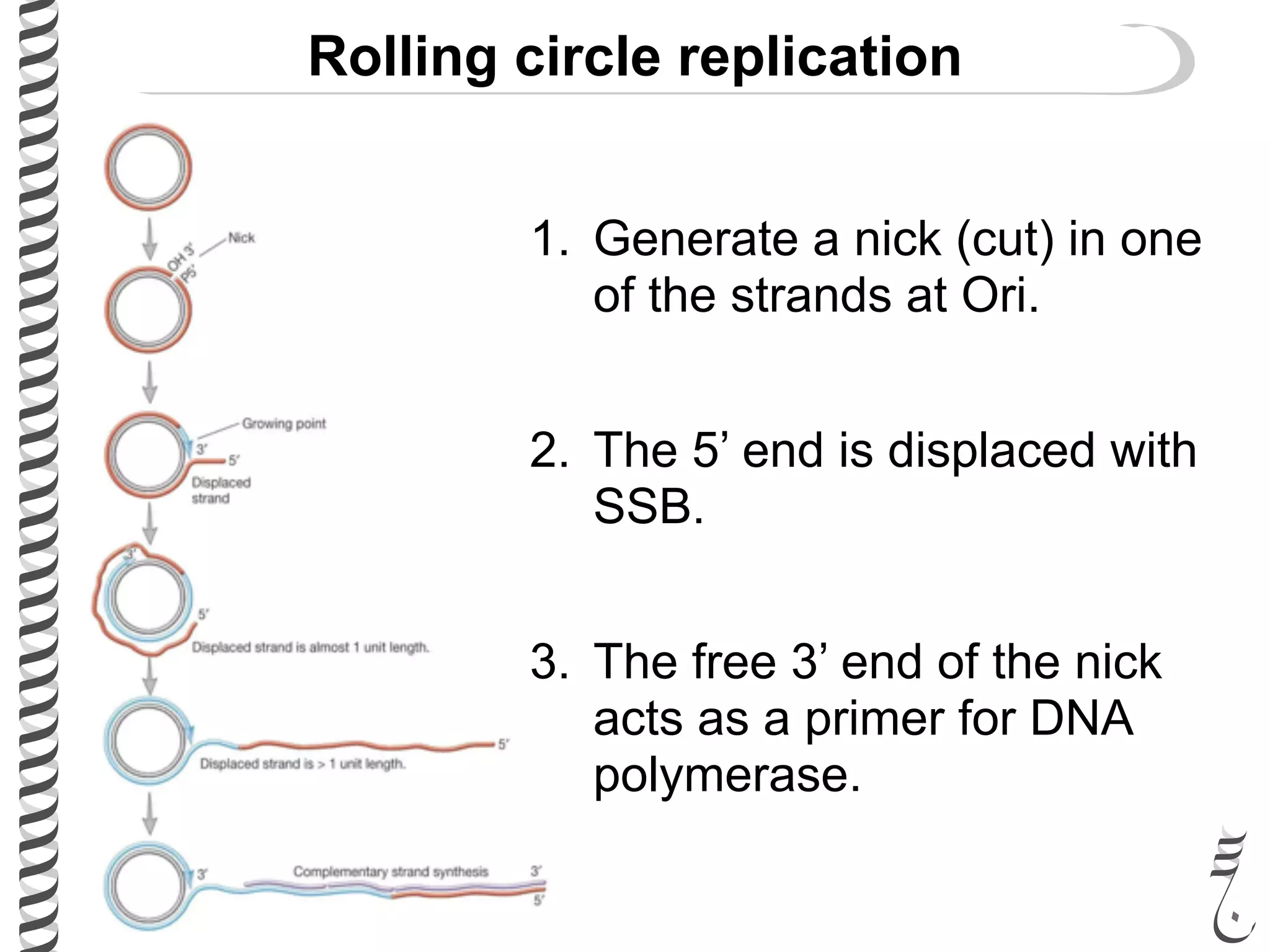
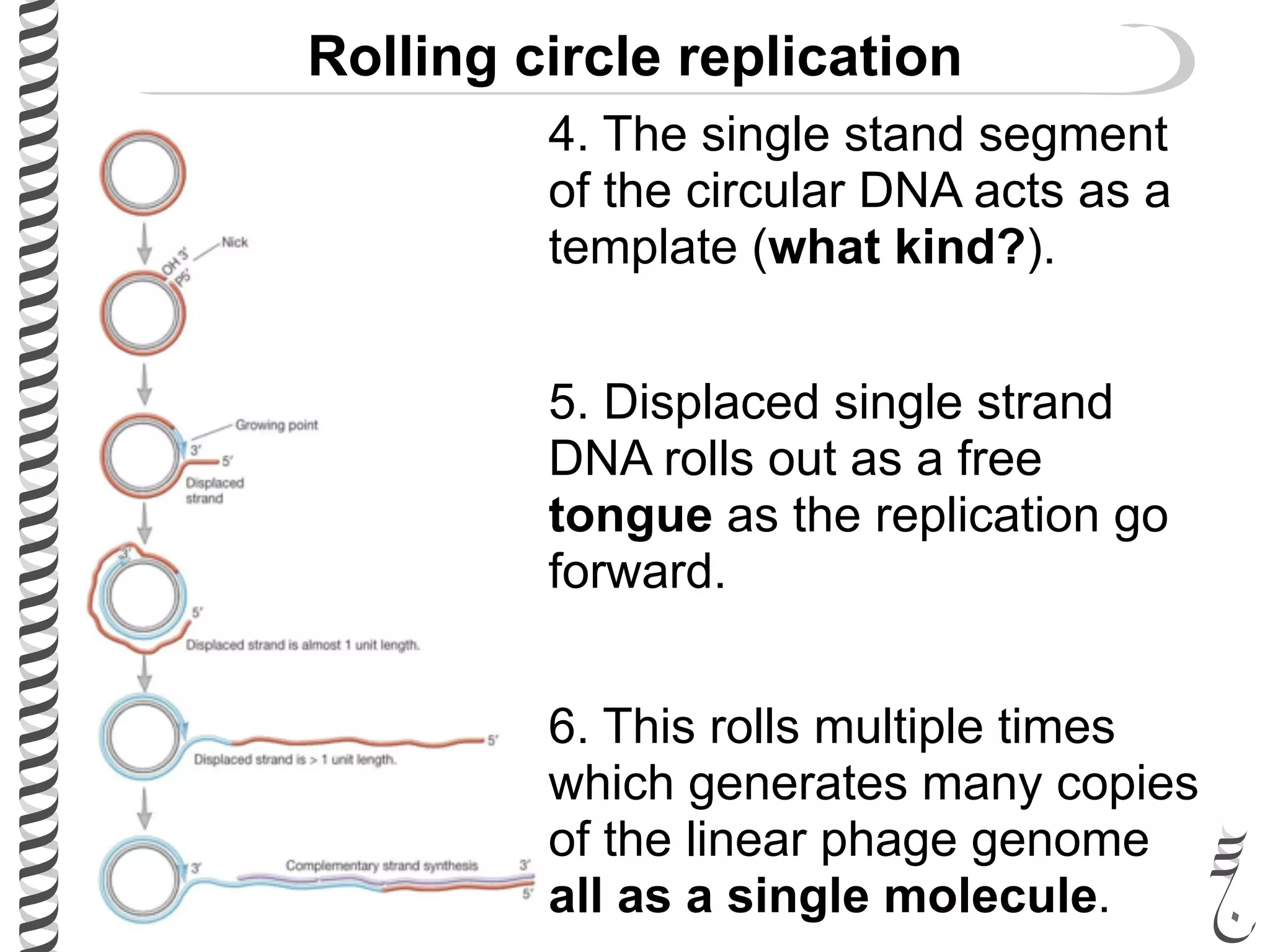
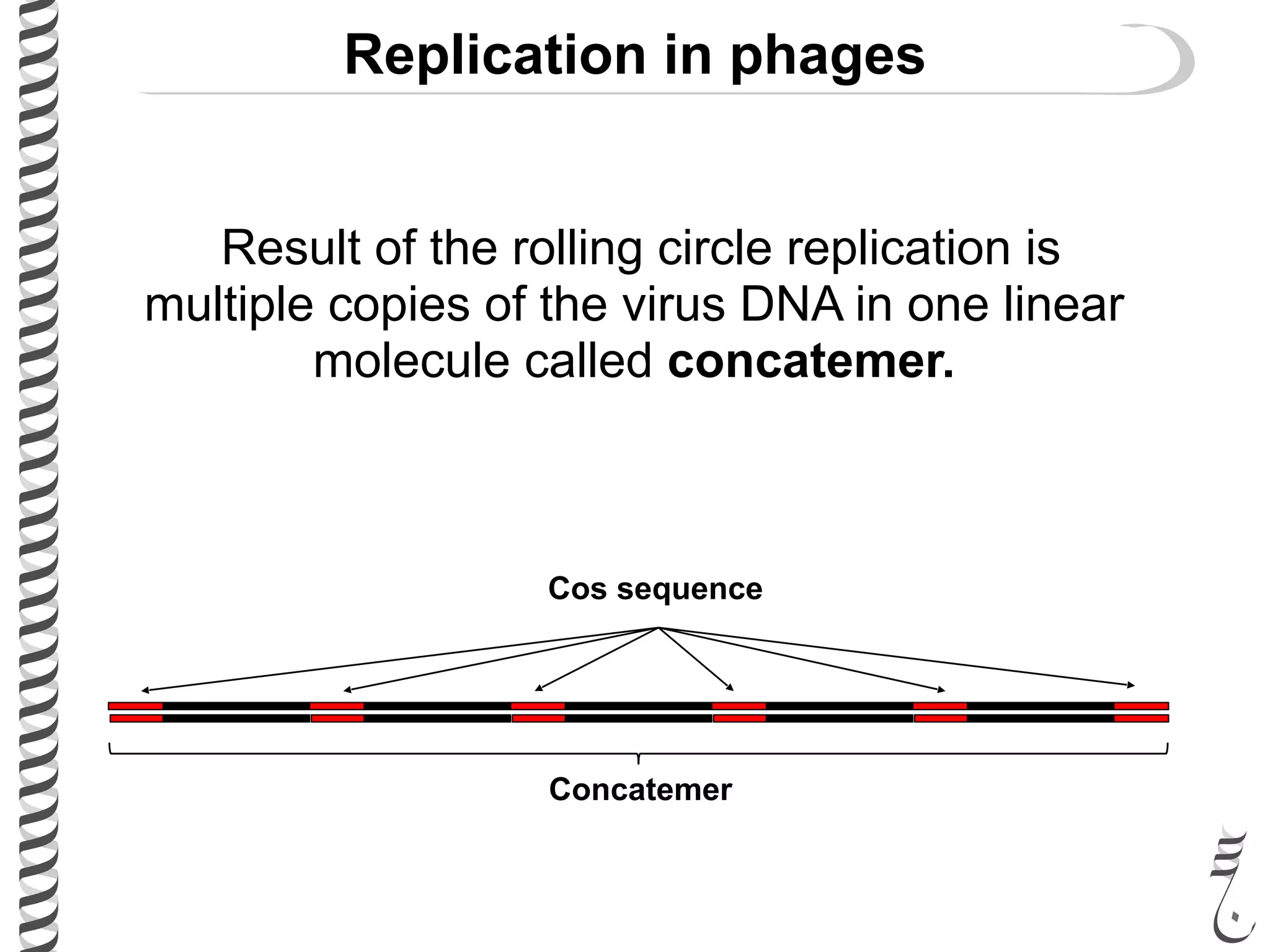
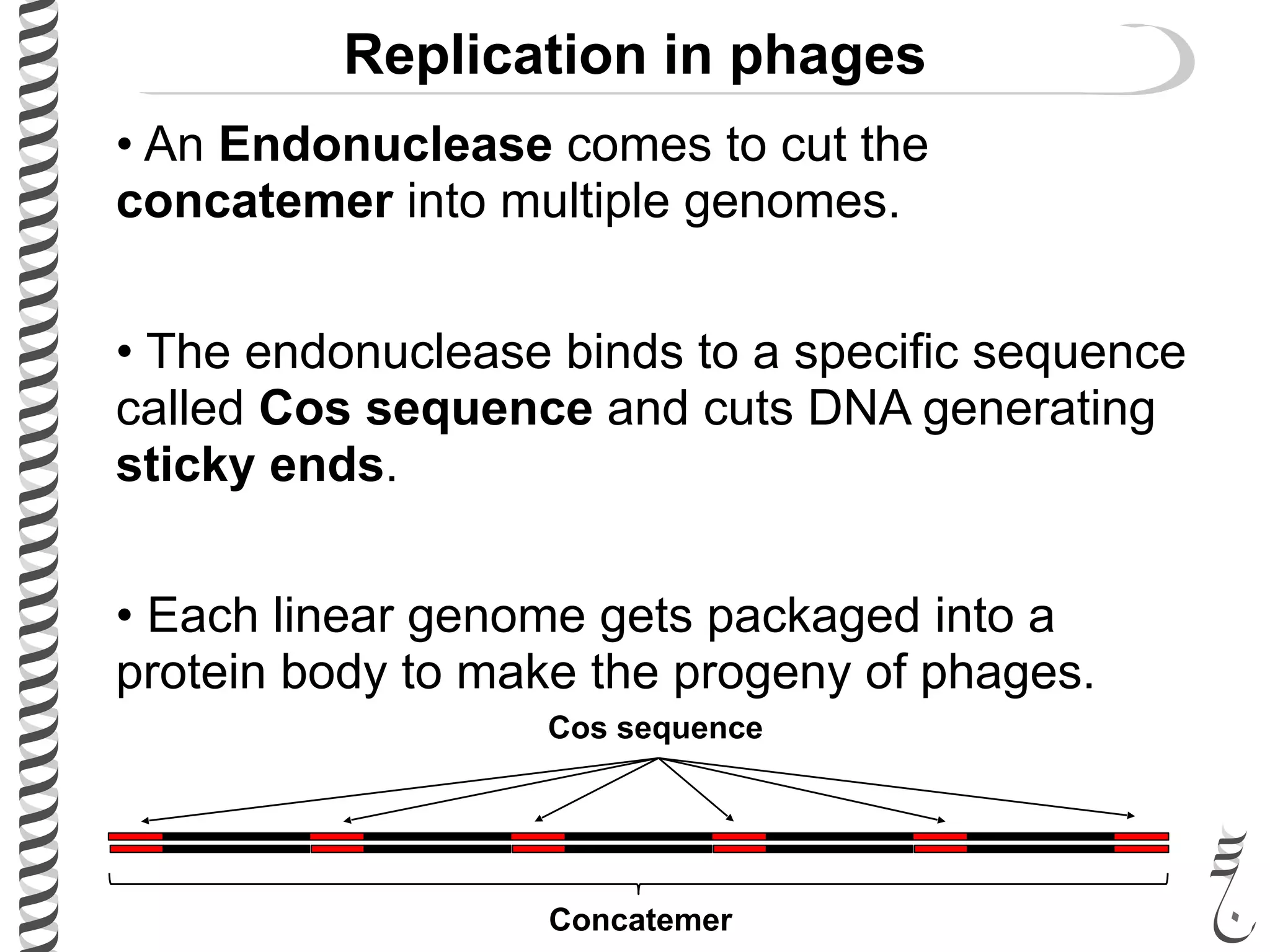
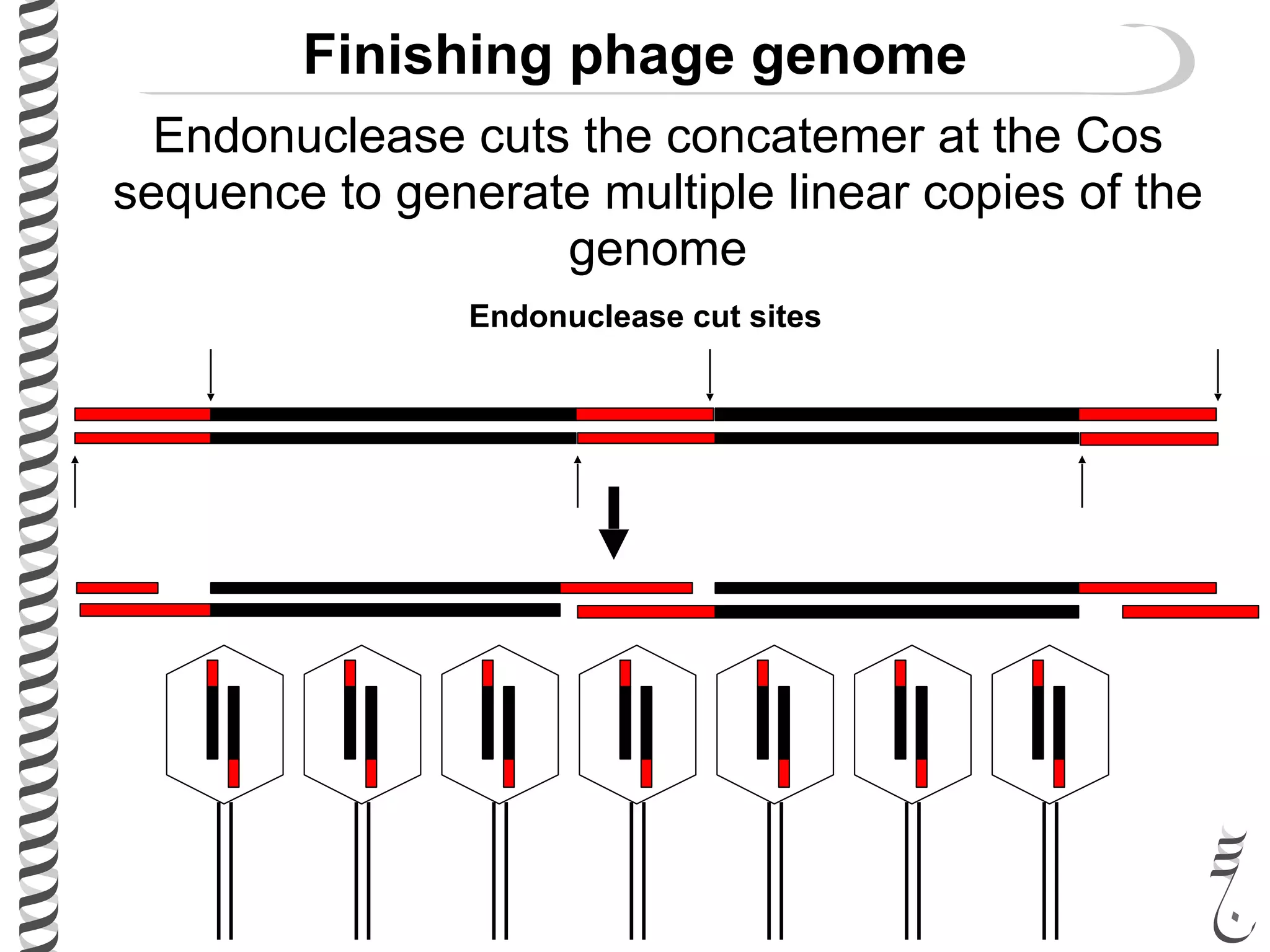
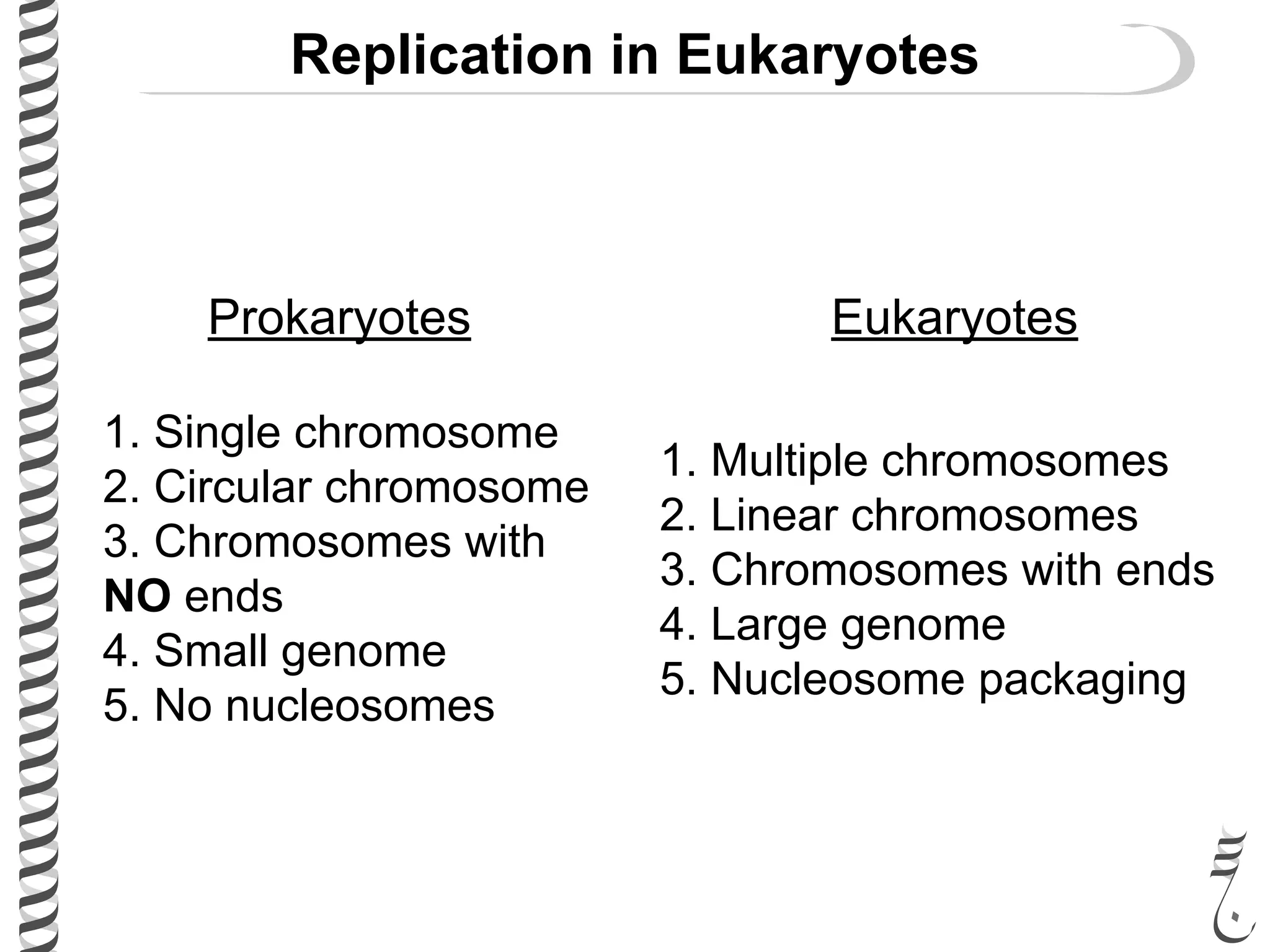
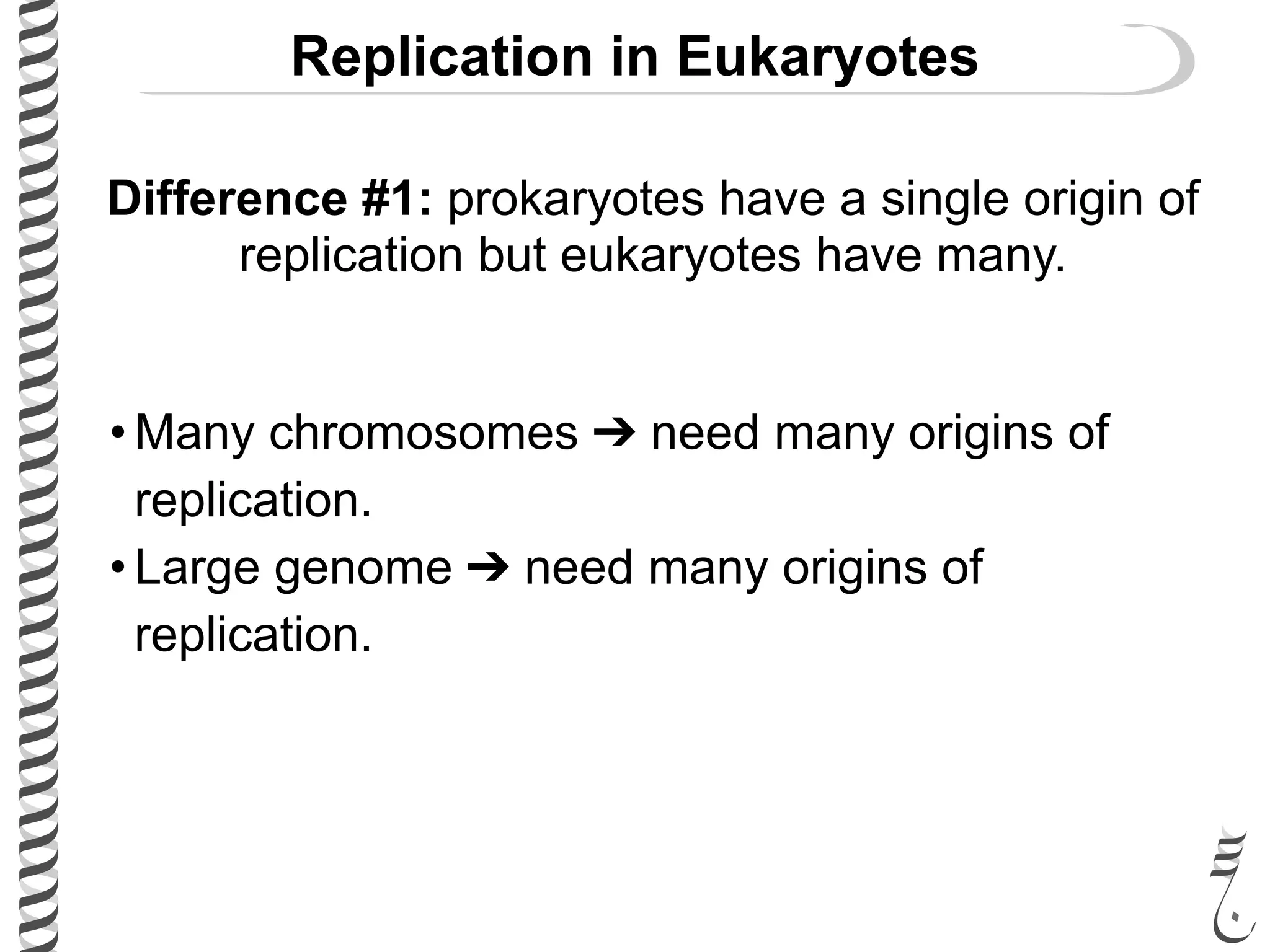
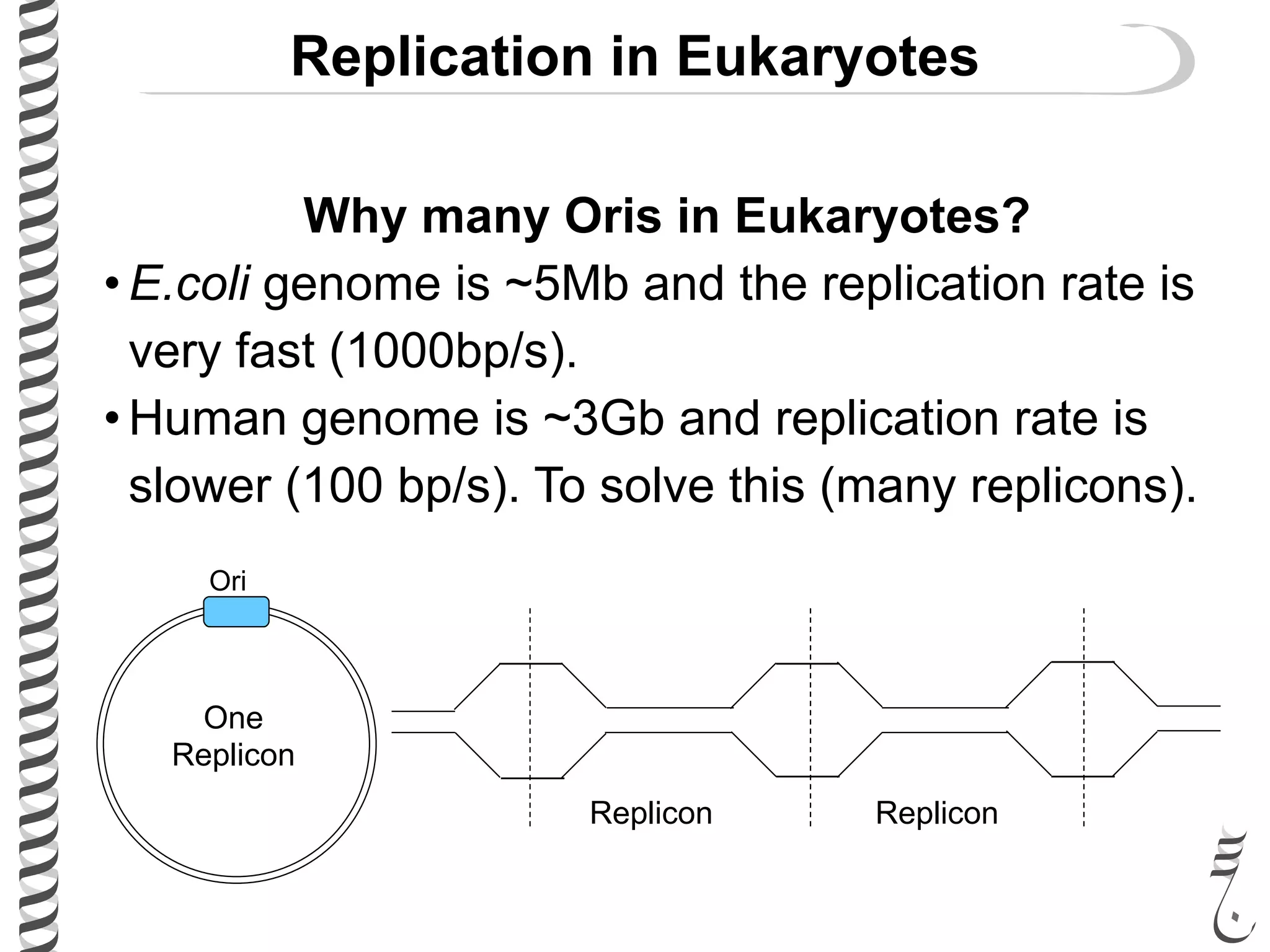
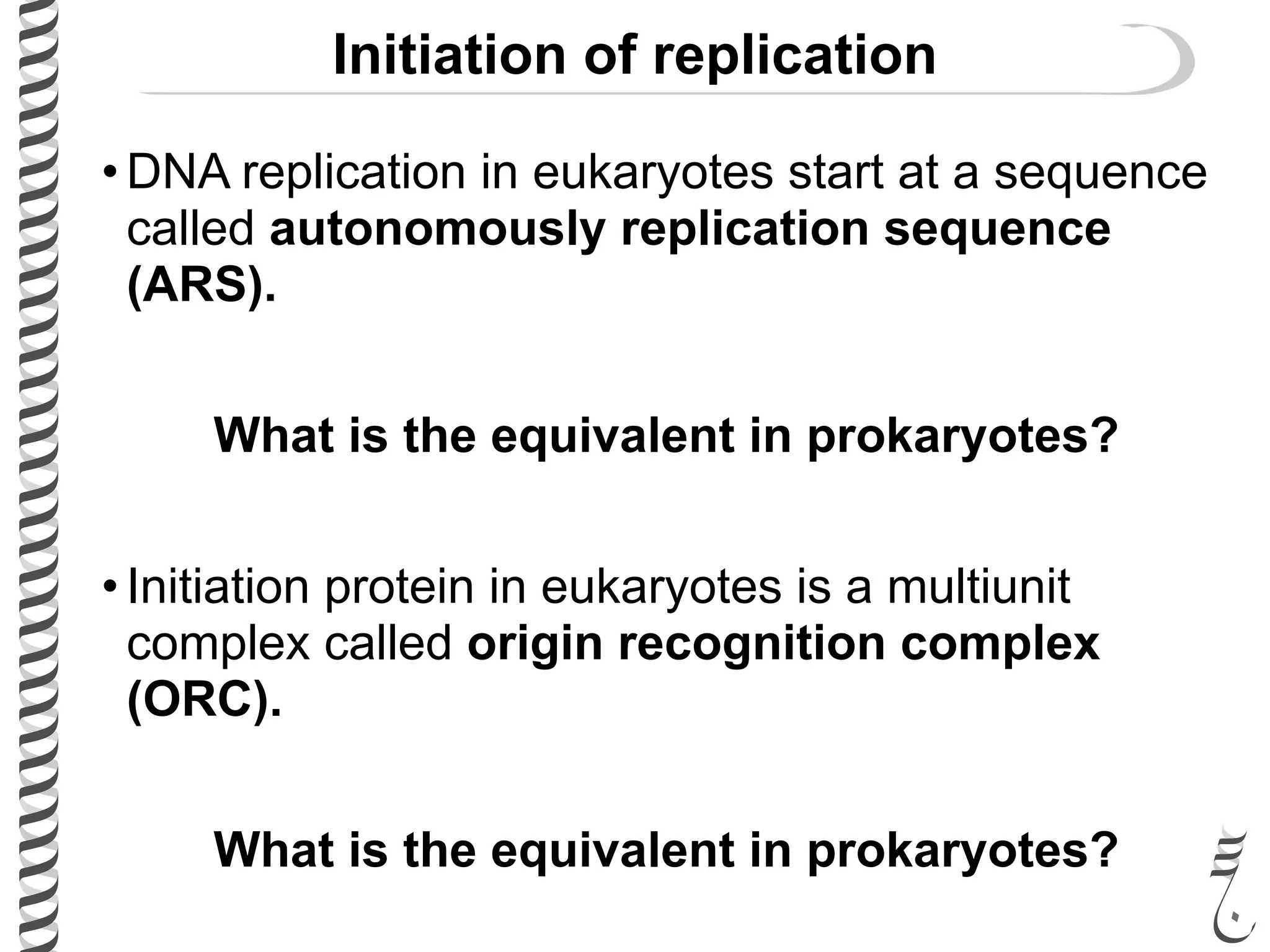
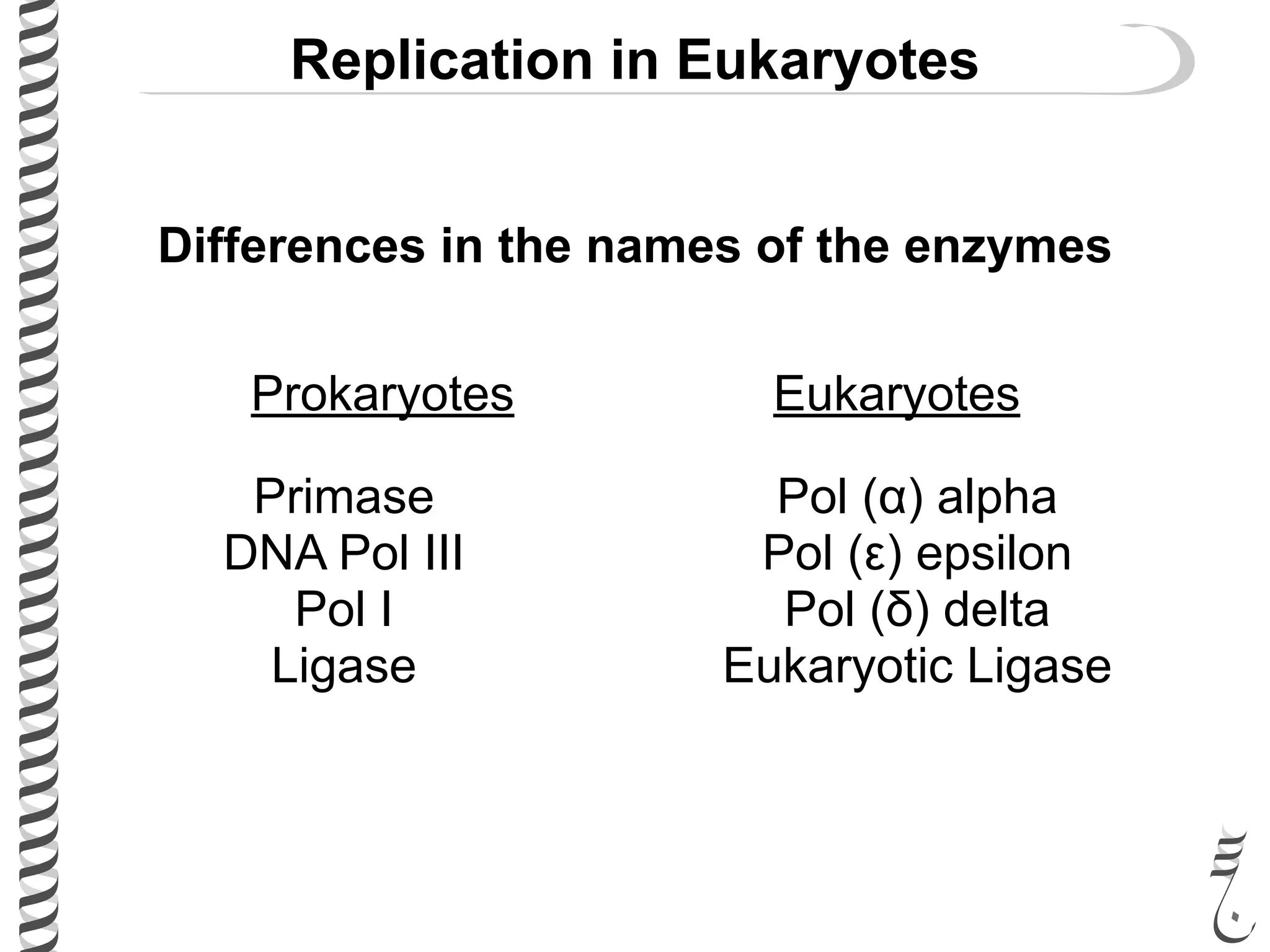
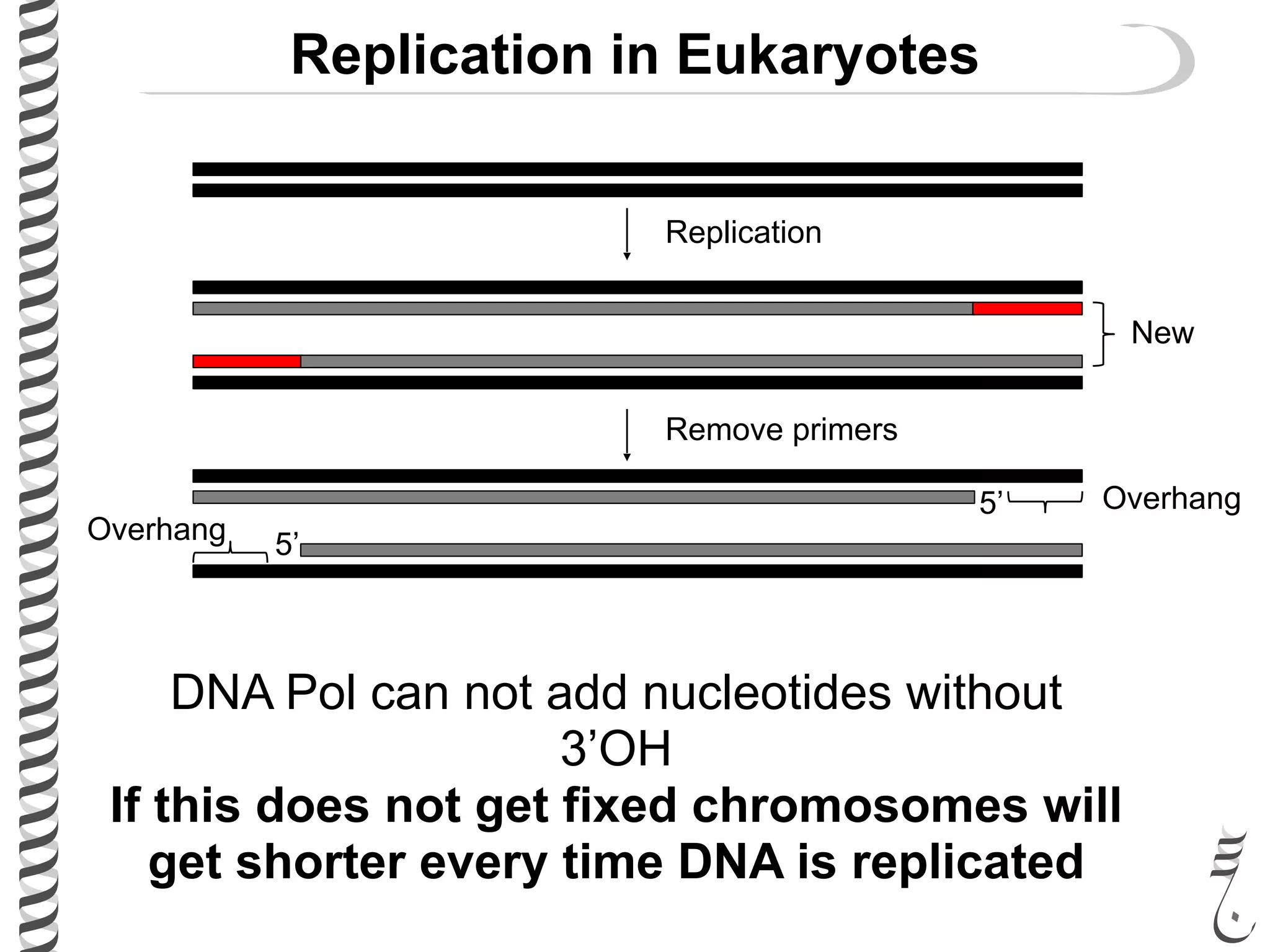
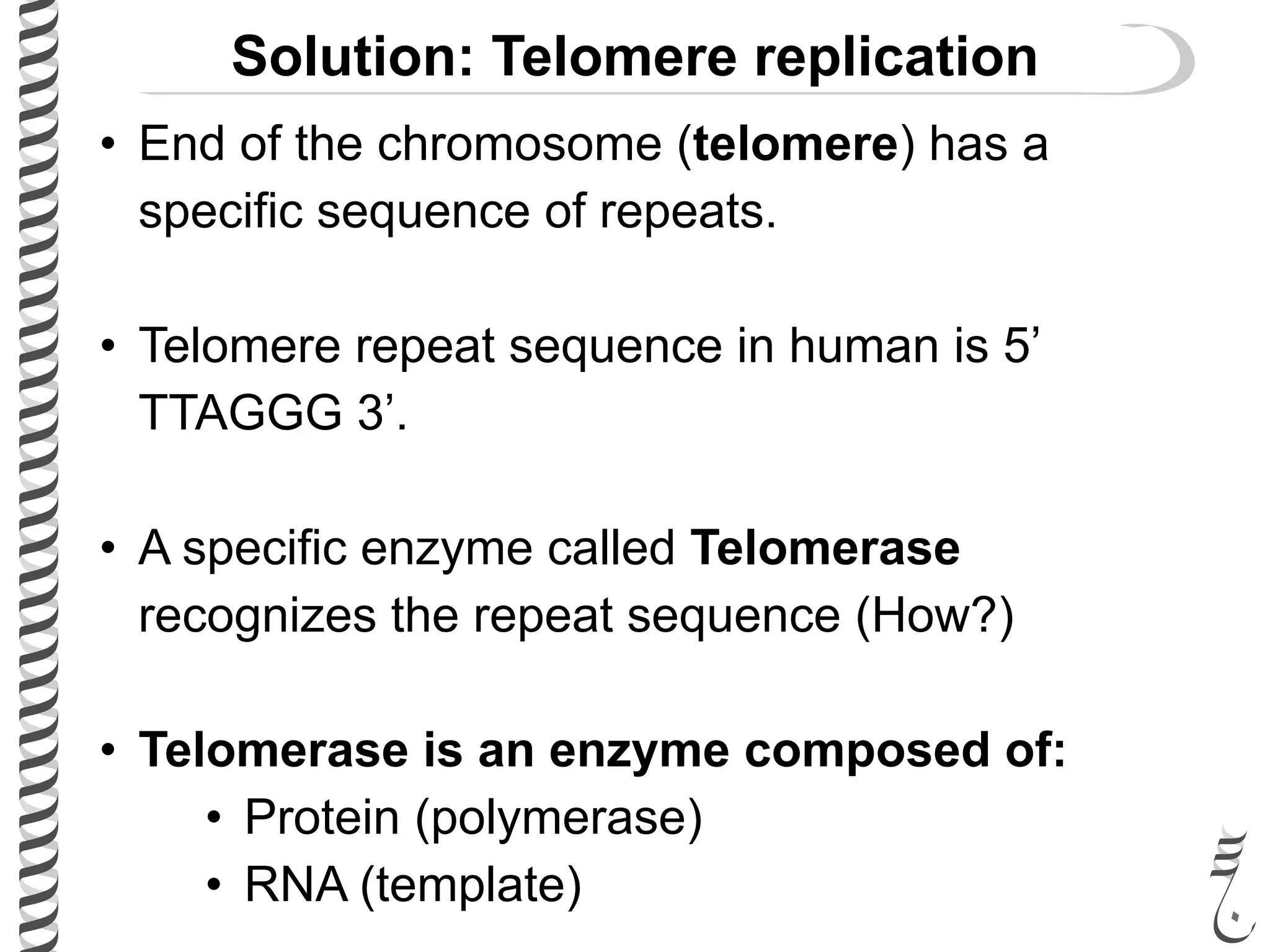
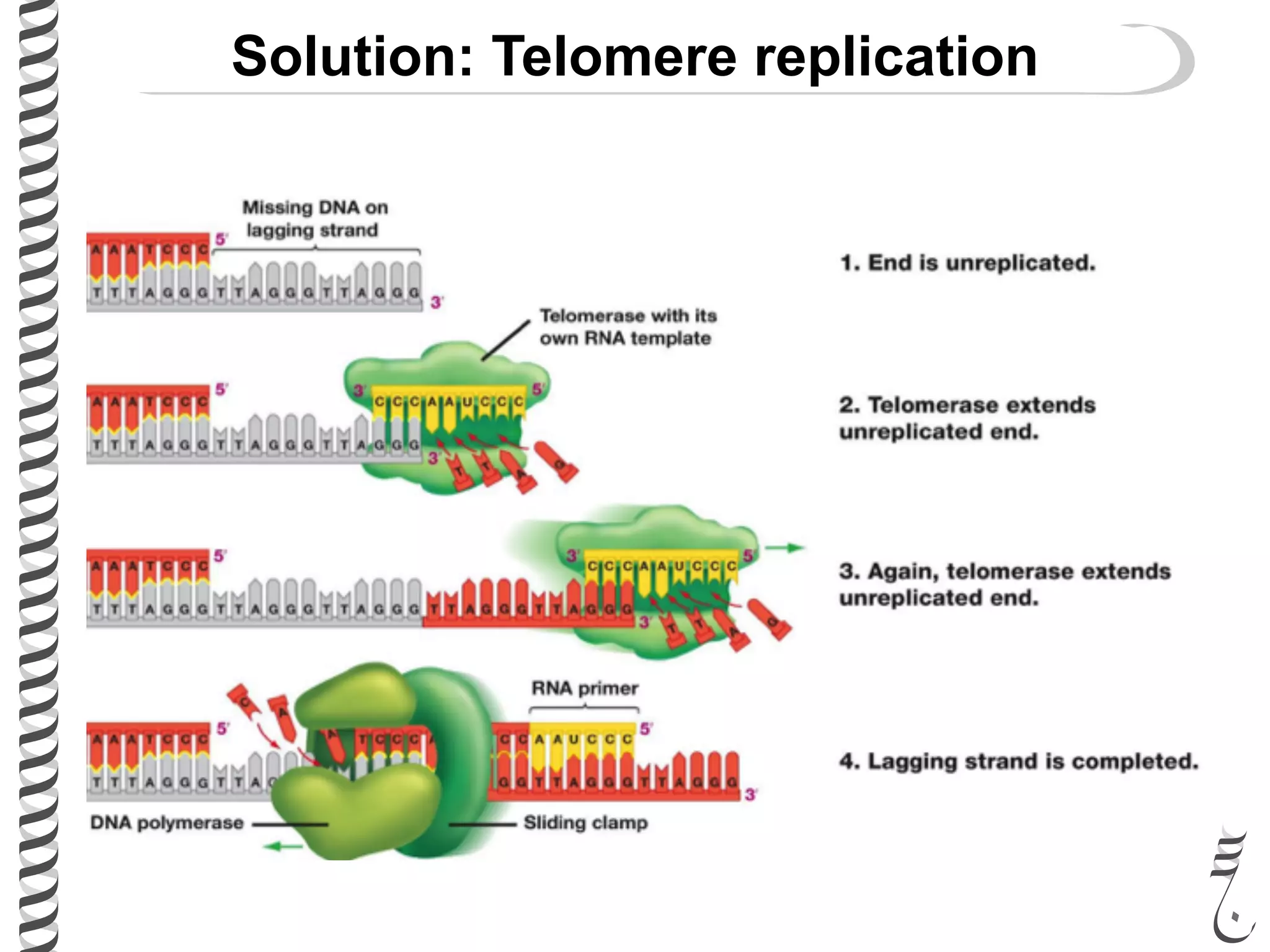
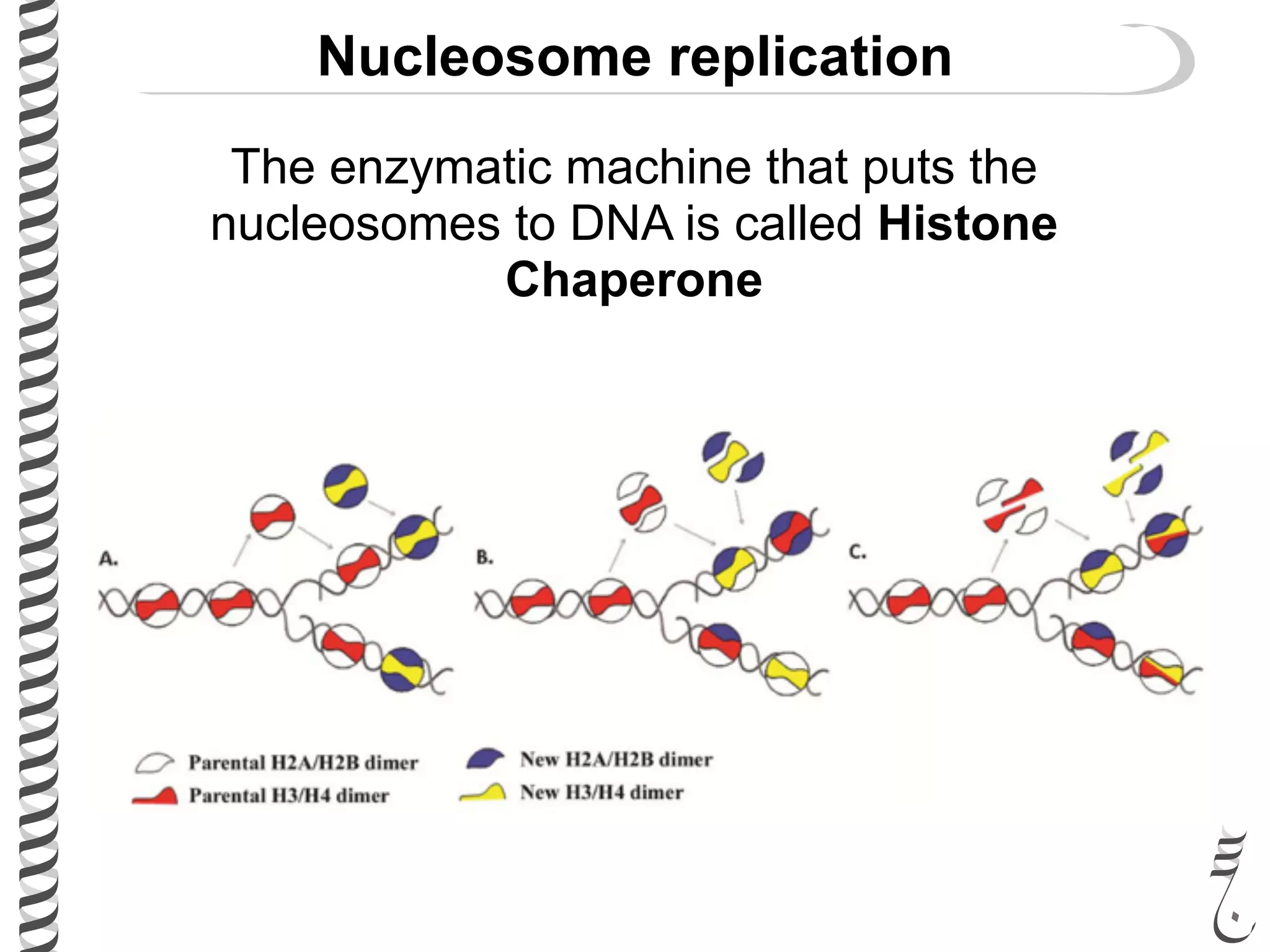
The document discusses DNA replication in bacteriophages and eukaryotes. In bacteriophages, rolling circle replication produces long concatemers that are then cleaved by an endonuclease into individual linear genomes. Eukaryotic replication differs from prokaryotes in that eukaryotes have multiple chromosomes, linear chromosomes with telomeres, and nucleosomes. Telomeres are replicated by the reverse transcription of an RNA template by telomerase. Nucleosomes are replicated through the addition of new histone proteins to accommodate the packaging of two genomes.