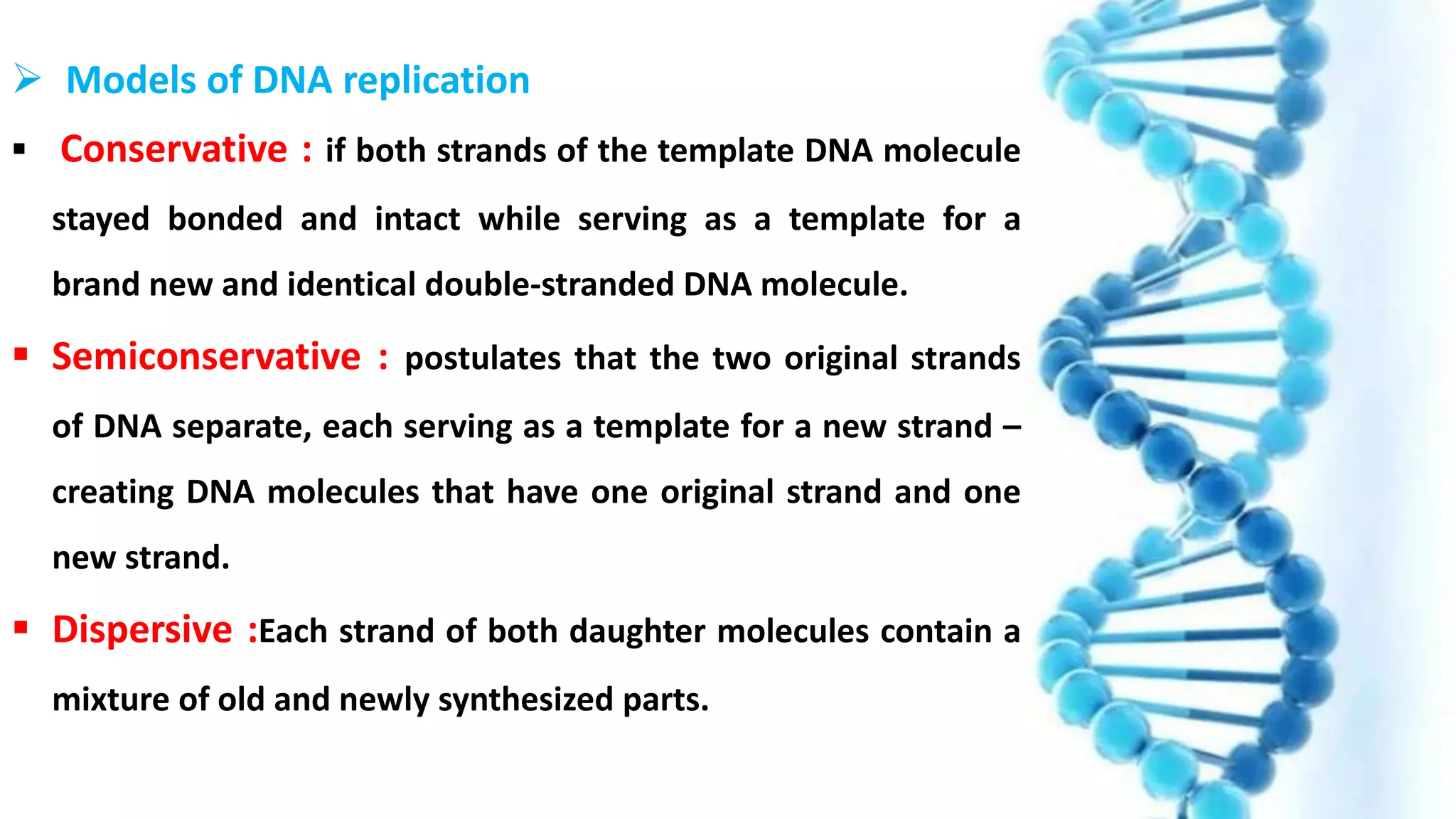
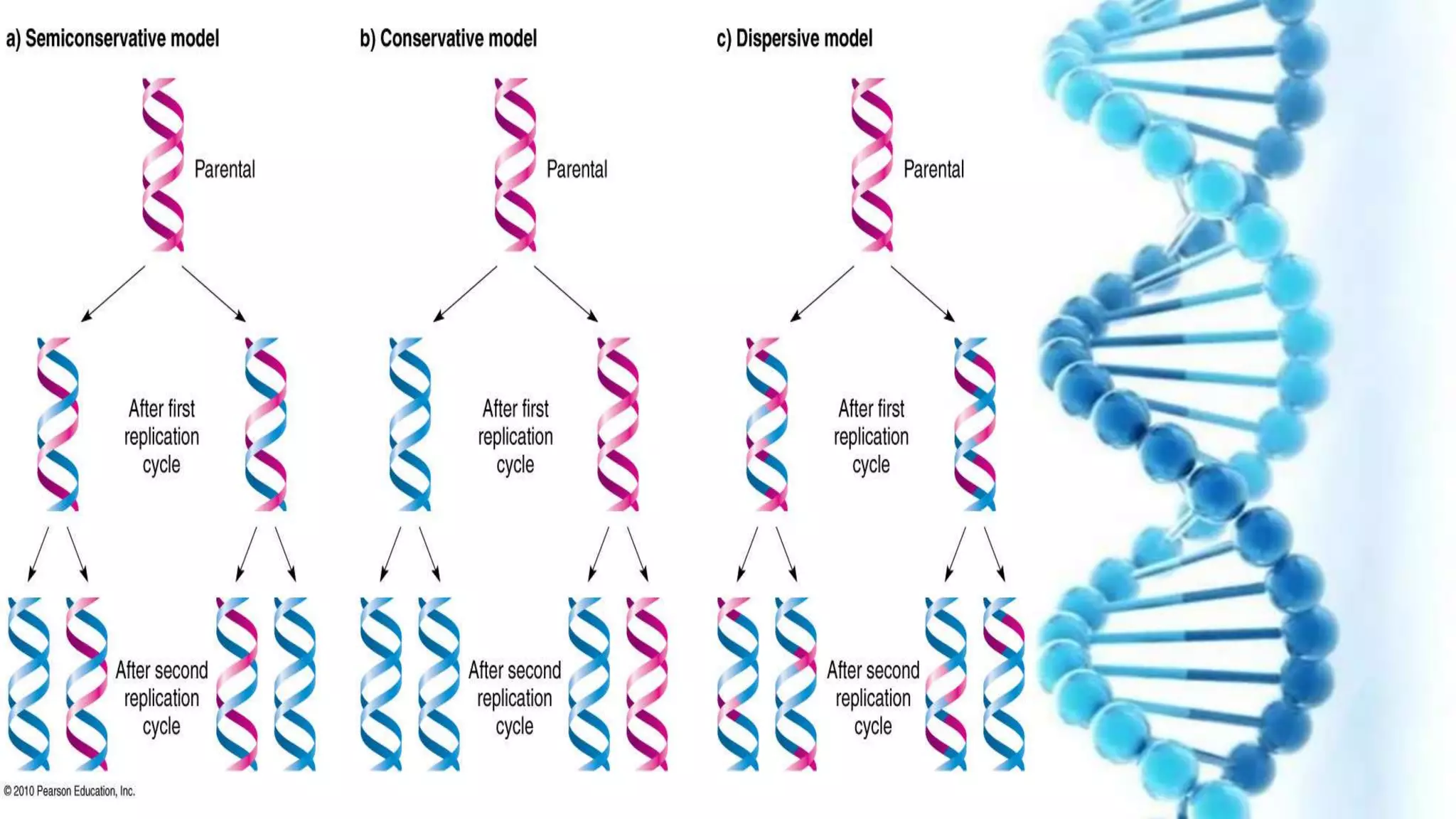
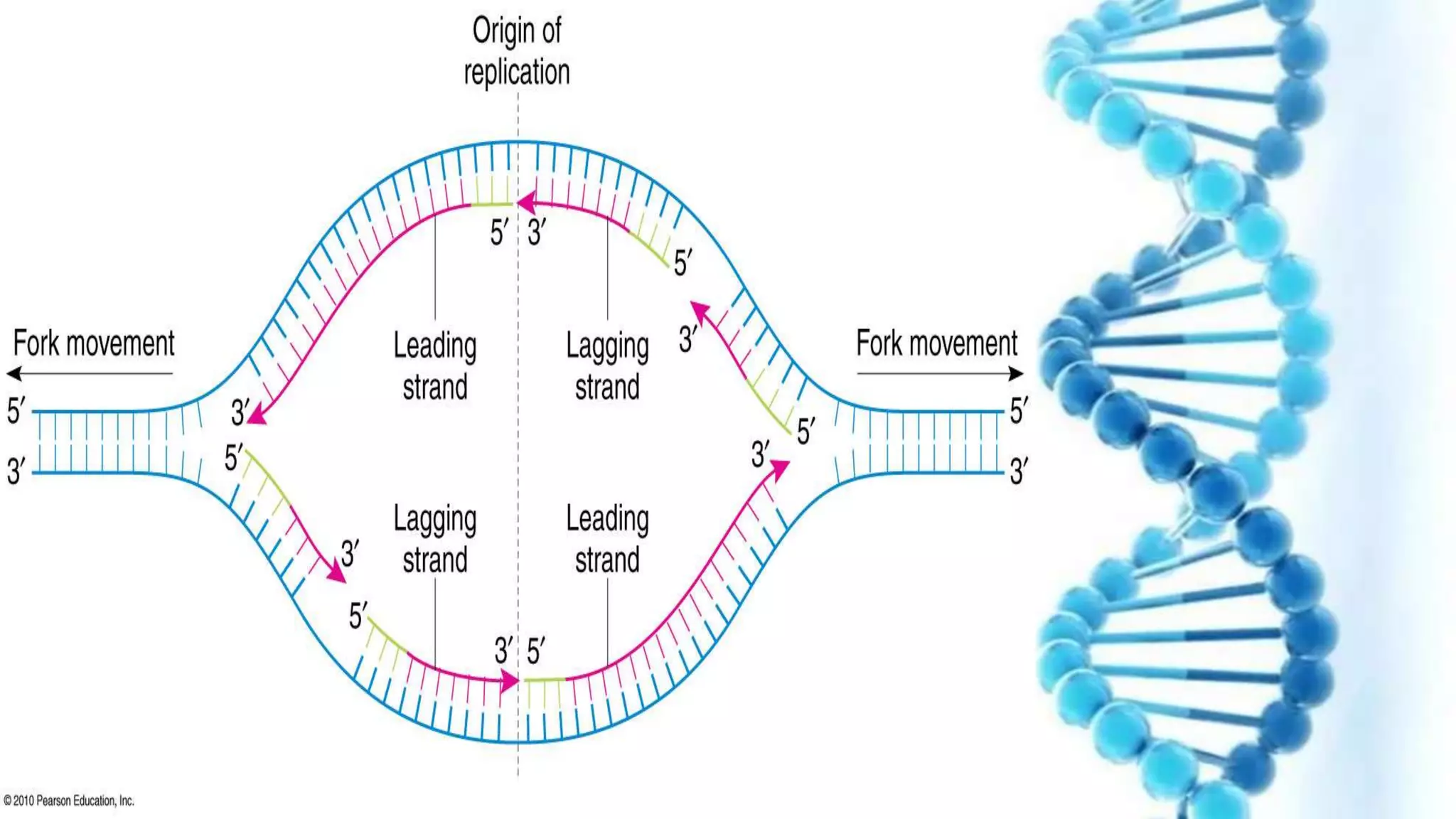
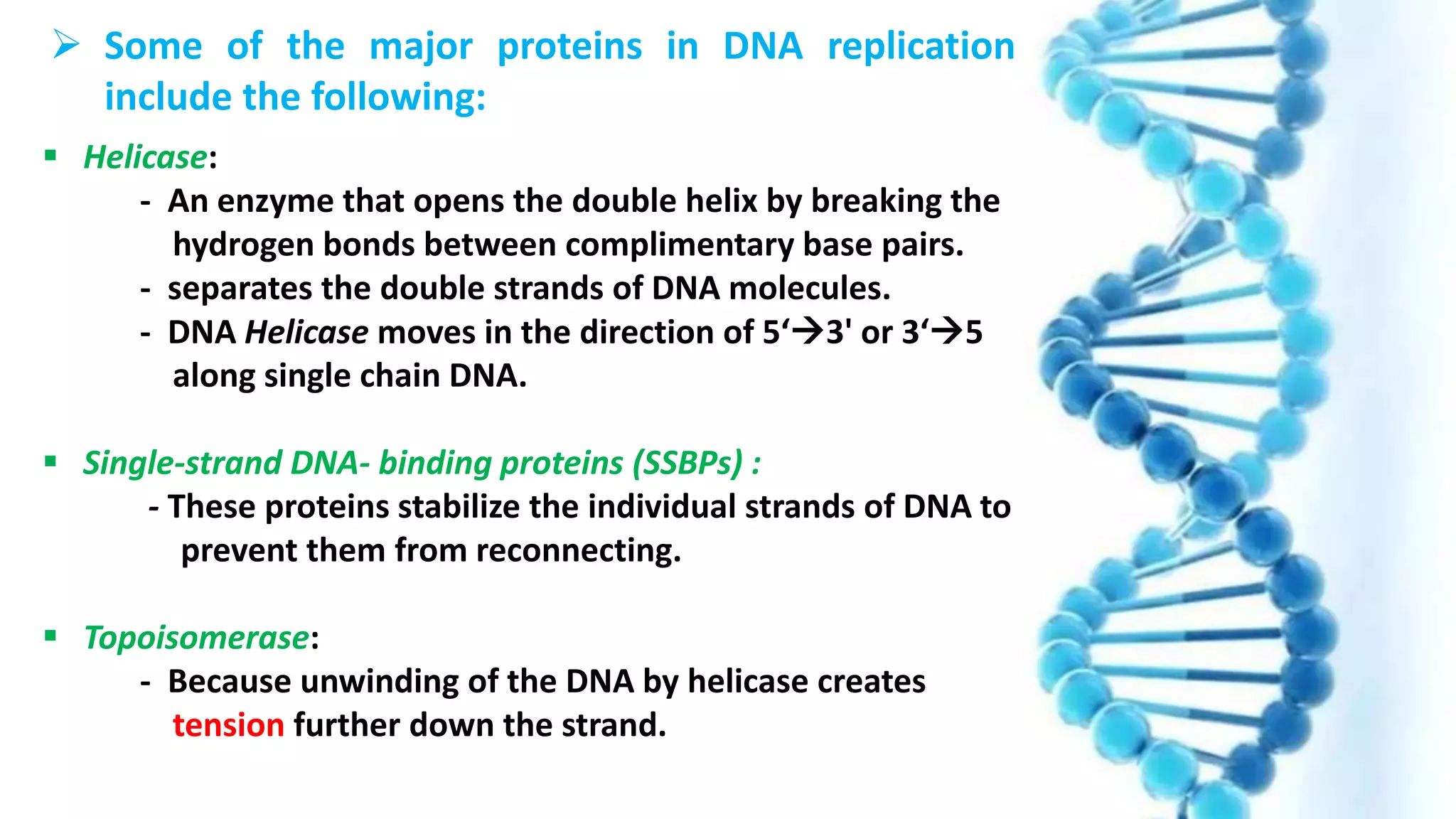
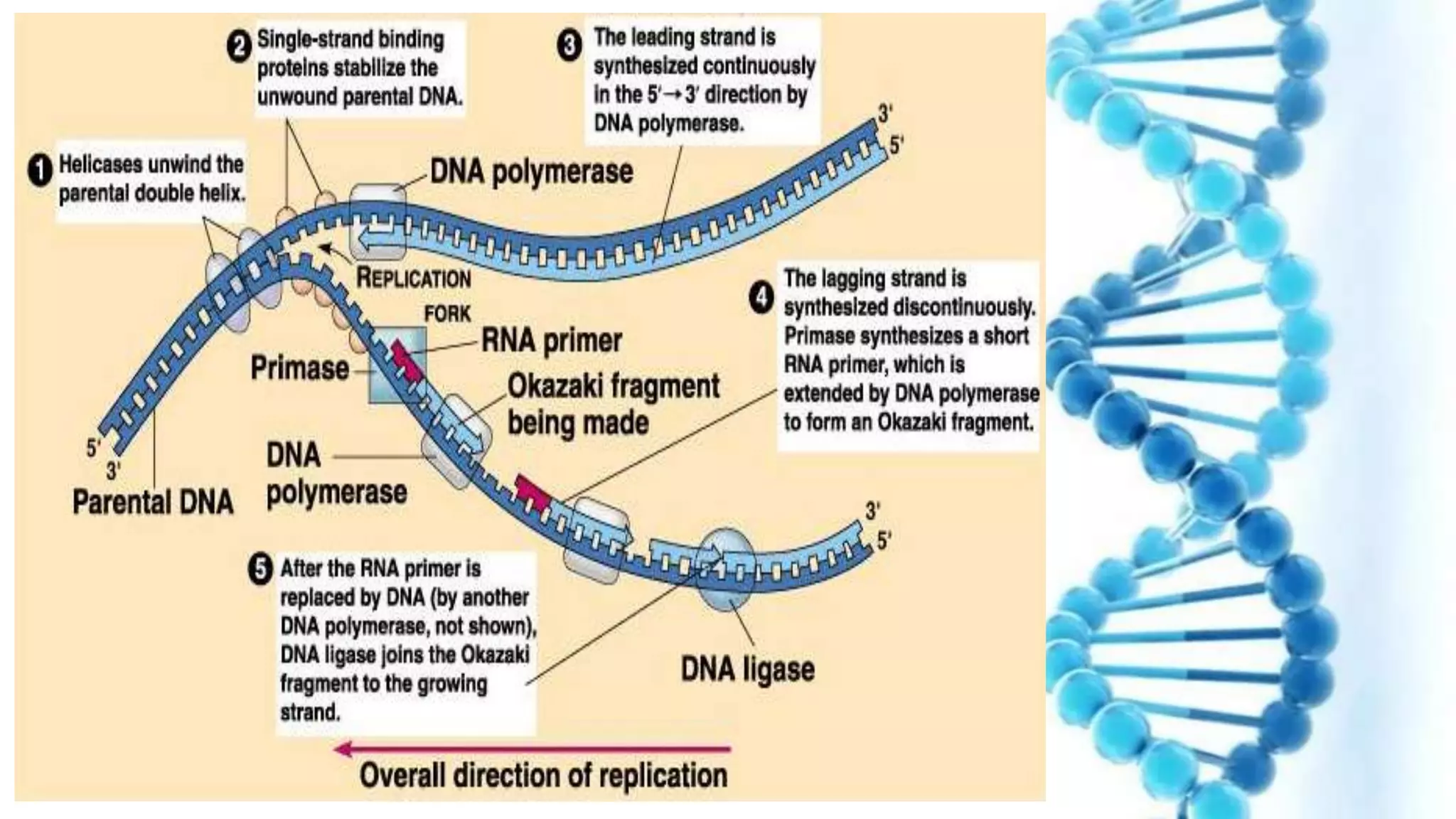
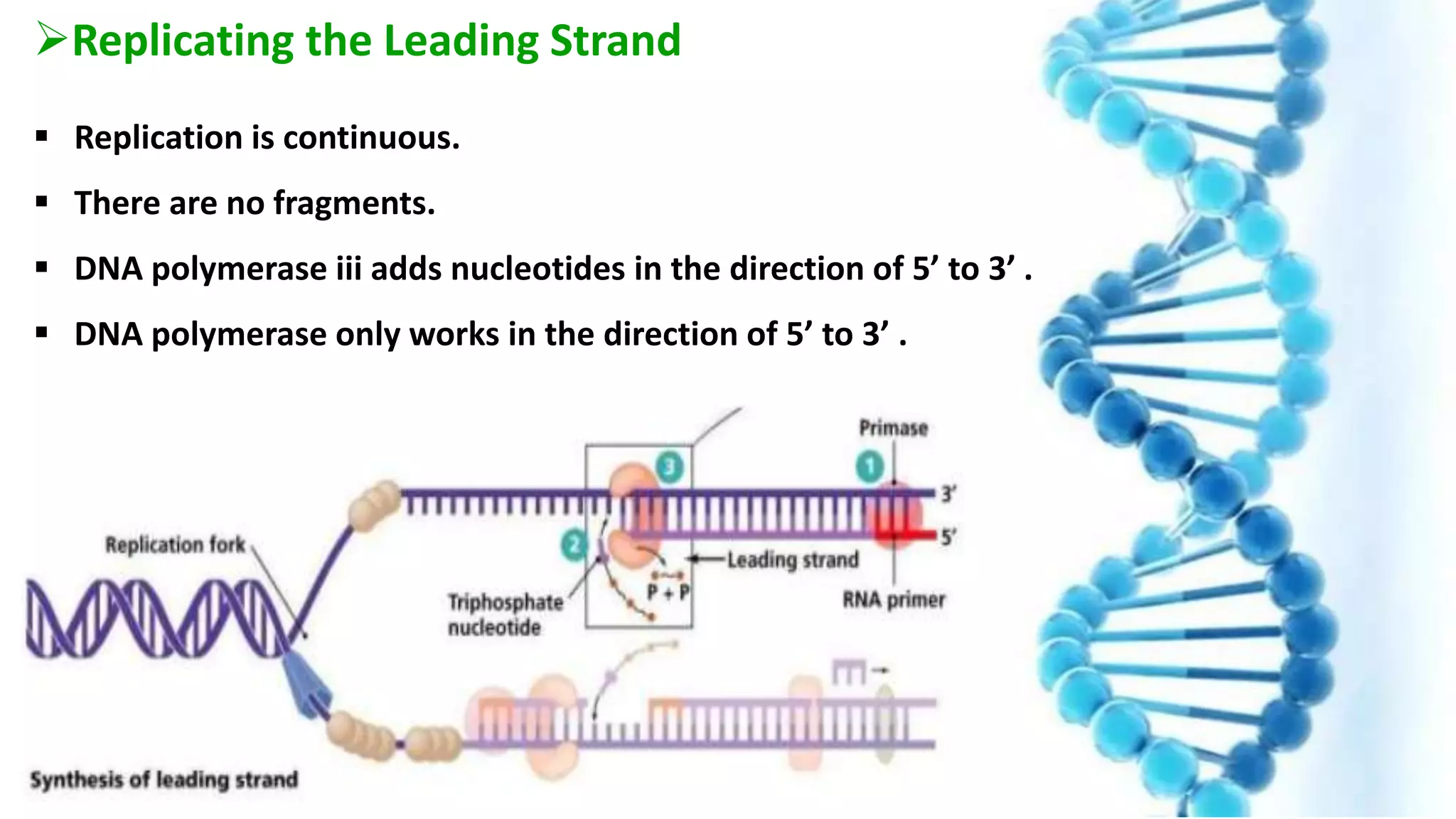
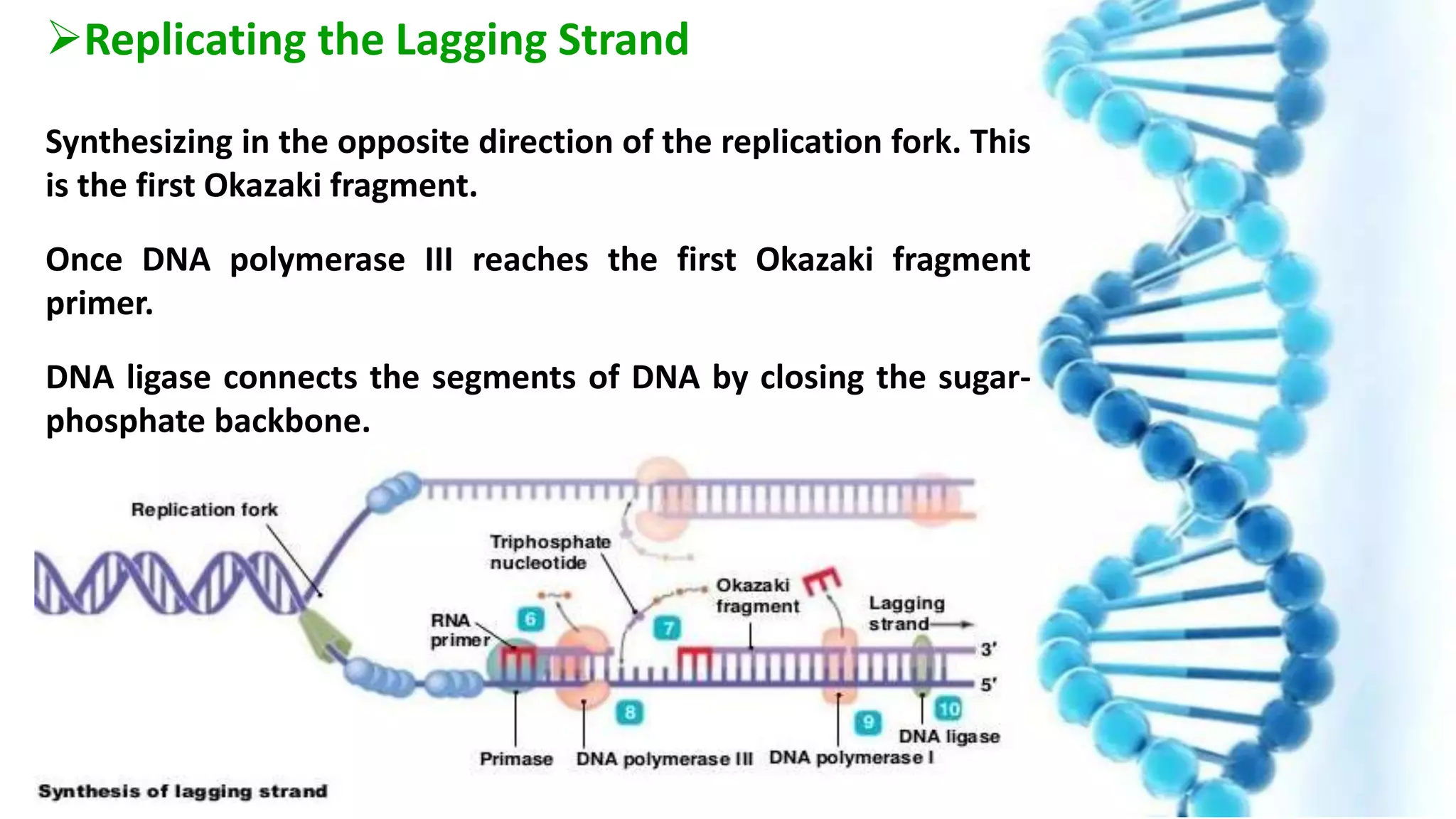
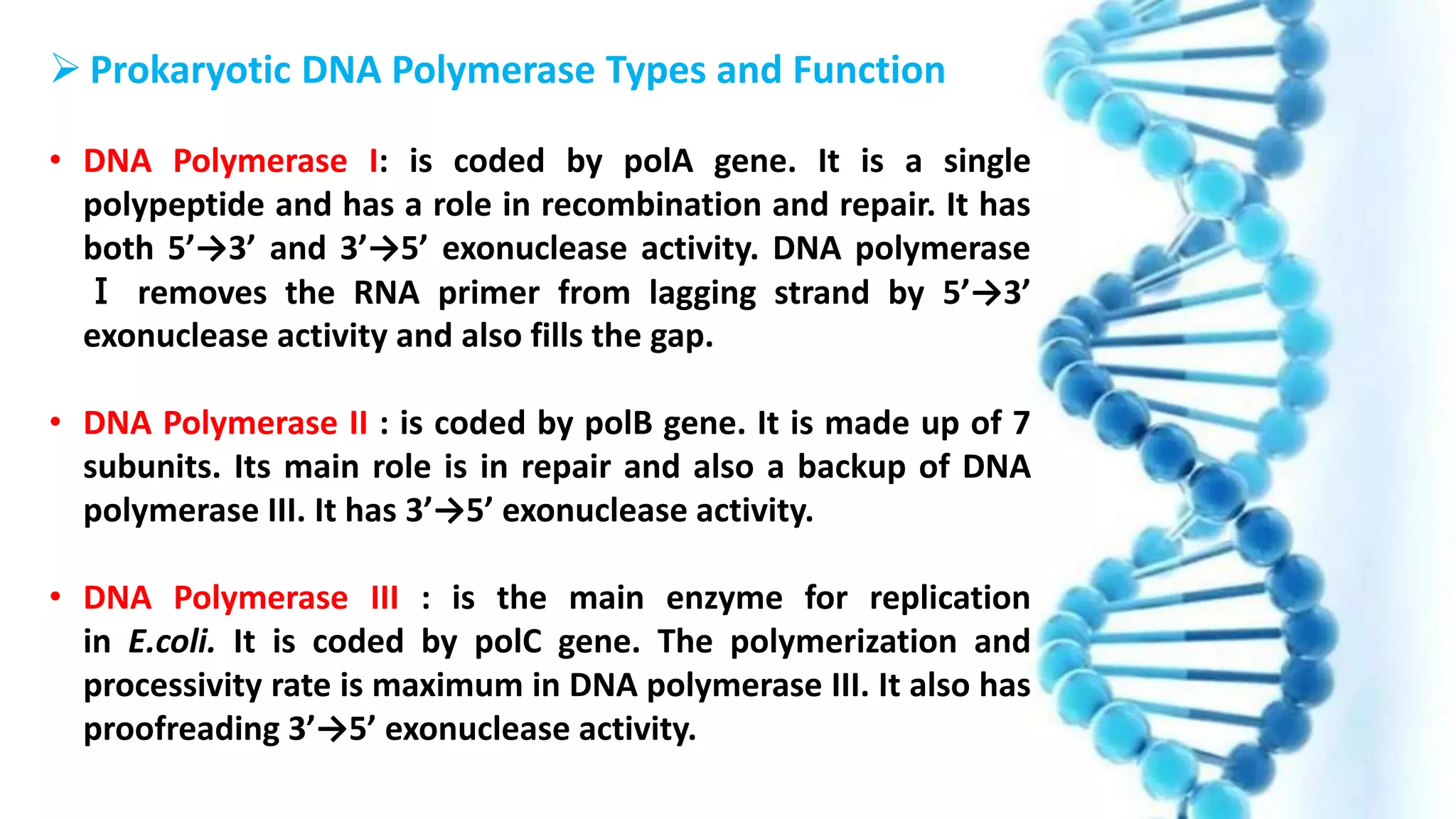
DNA replication is the process by which DNA copies itself in order to pass genetic information to daughter cells. It occurs through a semi-conservative process whereby the double helix unwinds and each strand acts as a template for new partner strands. DNA polymerase adds nucleotides to extend the new strands from primers in the 5' to 3' direction. On the leading strand synthesis is continuous but on the lagging strand it occurs discontinuously in fragments called Okazaki fragments that are later joined by DNA ligase. Telomerase adds repetitive sequences to the ends of lagging strands during replication to compensate for nucleotide losses and prevent shortening of chromosomes.