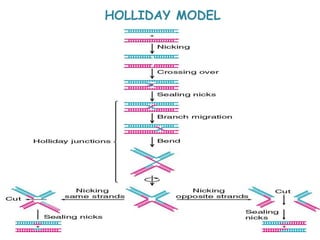
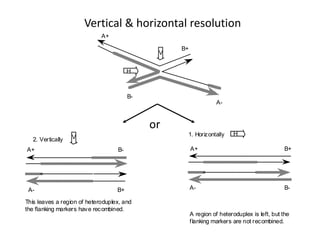
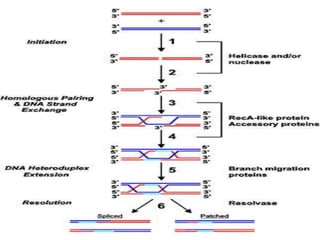
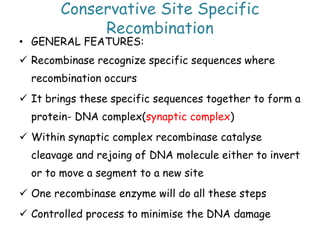
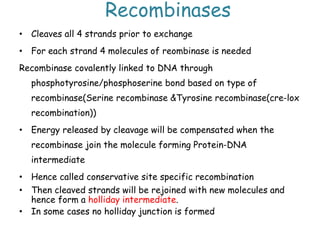
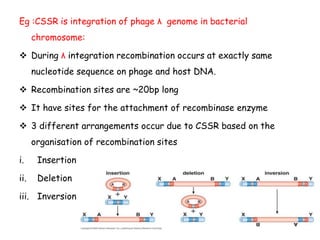
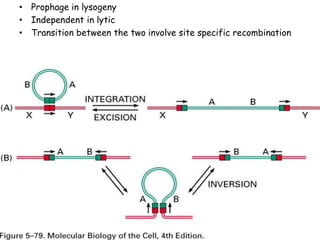
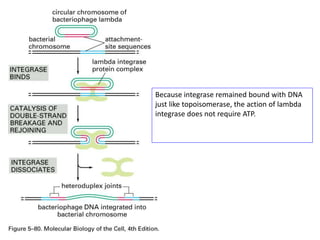
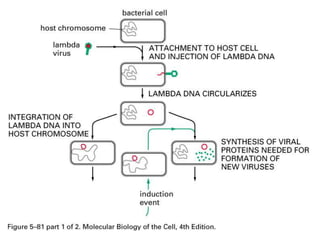
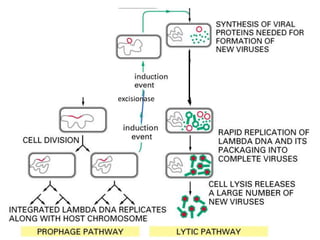
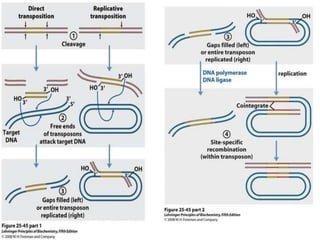
The document explains the molecular mechanisms of recombination, detailing the processes of homologous recombination, including crossing over, gene conversion, and various steps involved in forming Holliday junctions. It discusses different models of recombination such as the double-strand break model and site-specific recombination, highlighting the roles of specific proteins and enzymes. Additionally, it describes types of site-specific recombination, including conservative and transpositional types, emphasizing their applications in genetic engineering.